We can take a look at the design table to see the initial design.
d_model: 8, dim_feedforward: 16
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 88 │ train │ 20.5 K │ [128, 10] │ [128, 8] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [128, 8] │ [128, 128, 8] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 4.8 K │ train │ 134 M │ [128, 128, 8] │ [128, 128, 8] │
│ 3 │ fc_out │ Linear │ 9 │ train │ 2.0 K │ [128, 8] │ [128, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 4.9 K
Non-trainable params: 0
Total params: 4.9 K
Total estimated model params size (MB): 0
Modules in train mode: 85
Modules in eval mode: 0
Total FLOPs: 134 M
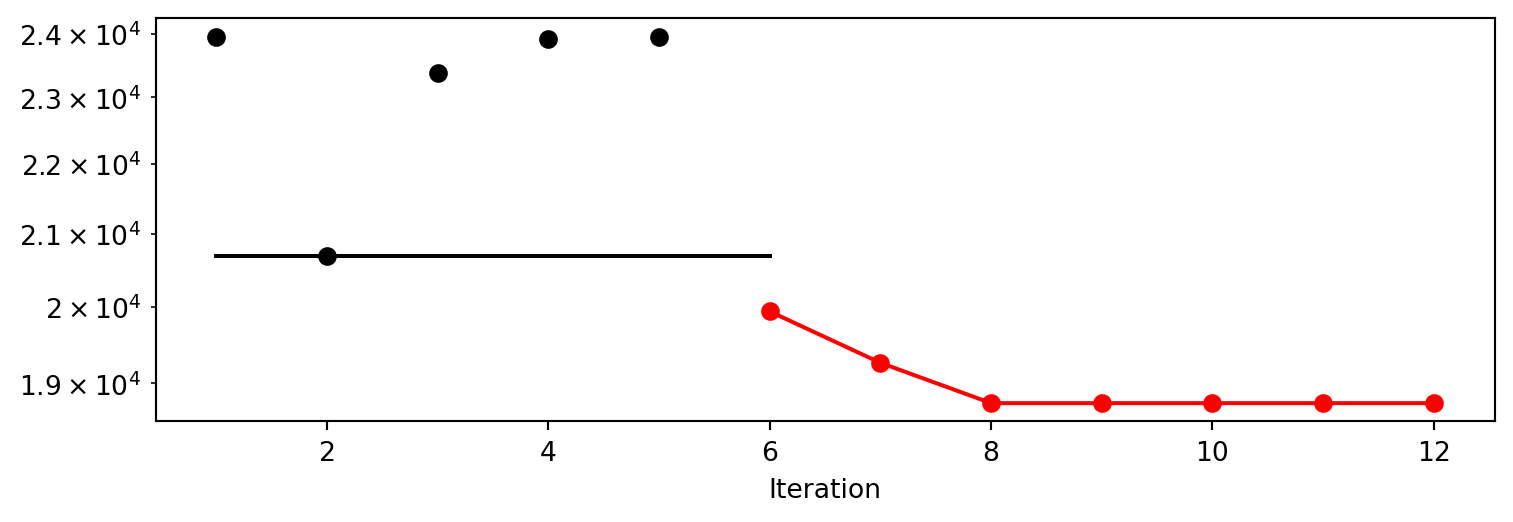
train_model result: {'val_loss': 23955.66015625, 'hp_metric': 23955.66015625}
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20858.498046875, 'hp_metric': 20858.498046875}
d_model: 32, dim_feedforward: 64
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 352 │ train │ 81.9 K │ [128, 10] │ [128, 32] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [128, 32] │ [128, 128, 32] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 68.4 K │ train │ 2.1 B │ [128, 128, 32] │ [128, 128, 32] │
│ 3 │ fc_out │ Linear │ 33 │ train │ 8.2 K │ [128, 32] │ [128, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴────────────────┴────────────────┘
Trainable params: 68.7 K
Non-trainable params: 0
Total params: 68.7 K
Total estimated model params size (MB): 0
Modules in train mode: 85
Modules in eval mode: 0
Total FLOPs: 2.1 B
train_model result: {'val_loss': 23231.283203125, 'hp_metric': 23231.283203125}
d_model: 16, dim_feedforward: 32
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 176 │ train │ 20.5 K │ [64, 10] │ [64, 16] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [64, 16] │ [64, 64, 16] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 35.6 K │ train │ 268 M │ [64, 64, 16] │ [64, 64, 16] │
│ 3 │ fc_out │ Linear │ 17 │ train │ 2.0 K │ [64, 16] │ [64, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴──────────────┴──────────────┘
Trainable params: 35.8 K
Non-trainable params: 0
Total params: 35.8 K
Total estimated model params size (MB): 0
Modules in train mode: 165
Modules in eval mode: 0
Total FLOPs: 268 M
train_model result: {'val_loss': 23903.546875, 'hp_metric': 23903.546875}
d_model: 8, dim_feedforward: 16
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 88 │ train │ 41.0 K │ [256, 10] │ [256, 8] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [256, 8] │ [256, 256, 8] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 1.2 K │ train │ 134 M │ [256, 256, 8] │ [256, 256, 8] │
│ 3 │ fc_out │ Linear │ 9 │ train │ 4.1 K │ [256, 8] │ [256, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 1.3 K
Non-trainable params: 0
Total params: 1.3 K
Total estimated model params size (MB): 0
Modules in train mode: 25
Modules in eval mode: 0
Total FLOPs: 134 M
train_model result: {'val_loss': 23954.796875, 'hp_metric': 23954.796875}
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20560.65234375, 'hp_metric': 20560.65234375}
spotpython tuning: 20560.65234375 [##--------] 19.03%. Success rate: 100.00%
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20688.98046875, 'hp_metric': 20688.98046875}
spotpython tuning: 20560.65234375 [####------] 38.12%. Success rate: 50.00%
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20638.8046875, 'hp_metric': 20638.8046875}
spotpython tuning: 20560.65234375 [######----] 56.95%. Success rate: 33.33%
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20515.283203125, 'hp_metric': 20515.283203125}
spotpython tuning: 20515.283203125 [########--] 75.97%. Success rate: 50.00%
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 264 K │ train │ 536 M │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 266 K
Non-trainable params: 0
Total params: 266 K
Total estimated model params size (MB): 1
Modules in train mode: 25
Modules in eval mode: 0
Total FLOPs: 536 M
train_model result: {'val_loss': 21880.328125, 'hp_metric': 21880.328125}
spotpython tuning: 20515.283203125 [#########-] 87.93%. Success rate: 40.00%
d_model: 128, dim_feedforward: 256
┏━━━┳━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━┳━━━━━━━┳━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
┃ ┃ Name ┃ Type ┃ Params ┃ Mode ┃ FLOPs ┃ In sizes ┃ Out sizes ┃
┡━━━╇━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━╇━━━━━━━╇━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
│ 0 │ input_proj │ Linear │ 1.4 K │ train │ 81.9 K │ [32, 10] │ [32, 128] │
│ 1 │ positional_encoding │ PositionalEncoding │ 0 │ train │ 0 │ [32, 128] │ [32, 32, 128] │
│ 2 │ transformer_encoder │ TransformerEncoder │ 529 K │ train │ 1.1 B │ [32, 32, 128] │ [32, 32, 128] │
│ 3 │ fc_out │ Linear │ 129 │ train │ 8.2 K │ [32, 128] │ [32, 1] │
└───┴─────────────────────┴────────────────────┴────────┴───────┴────────┴───────────────┴───────────────┘
Trainable params: 531 K
Non-trainable params: 0
Total params: 531 K
Total estimated model params size (MB): 2
Modules in train mode: 45
Modules in eval mode: 0
Total FLOPs: 1.1 B
train_model result: {'val_loss': 20147.291015625, 'hp_metric': 20147.291015625}
spotpython tuning: 20147.291015625 [##########] 100.00%. Success rate: 50.00% Done...
Experiment saved to 603_res.pkl
Note that we have enabled Tensorboard-Logging, so we can visualize the results with Tensorboard. Execute the following command in the terminal to start Tensorboard.