import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from spotoptim import SpotOptim
from spotoptim.surrogate import Kriging
import time
import warnings
warnings.filterwarnings('ignore')
# Scikit-learn models
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import (
Matern, RBF, ConstantKernel, WhiteKernel, RationalQuadratic
)
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.svm import SVR16 Surrogate Model Selection in SpotOptim
This section demonstrates how to select and configure different surrogate models in SpotOptim. We compare various surrogate options:
- Gaussian Process with different kernels (Matern, RBF, Rational Quadratic)
- SpotOptim Kriging model
- Random Forest regressor
- XGBoost regressor
- Support Vector Regression (SVR)
- Gradient Boosting regressor
All methods are evaluated on the Aircraft Wing Weight Example (AWWE) function. We visualize the fitted surrogates and compare optimization performance.
16.1 Introduction
SpotOptim supports any scikit-learn compatible regressor, allowing you to choose the surrogate that best fits your problem characteristics:
- Gaussian Processes: Provide uncertainty estimates, smooth interpolation, good for continuous functions. Support Expected Improvement (EI) acquisition.
- Kriging: Similar to GP but with customizable correlation functions. Supports EI acquisition.
- Random Forests: Robust to noise, handle discontinuities. Don’t provide uncertainty, so use
acquisition='y'(greedy). - XGBoost: Excellent for high-dimensional problems, fast training and prediction. Use
acquisition='y'. - SVR: Good for high-dimensional problems with smooth structure. Use
acquisition='y'. - Gradient Boosting: Strong performance on structured problems. Use
acquisition='y'.
16.1.1 Acquisition Functions and Uncertainty
Models that provide uncertainty estimates (Gaussian Process, Kriging) work with all acquisition functions: ‘ei’ (Expected Improvement), ‘pi’ (Probability of Improvement), and ‘y’ (greedy).
Tree-based and other models (Random Forest, XGBoost, SVR, Gradient Boosting) don’t provide uncertainty estimates by default, so they should use acquisition='y' for greedy optimization. SpotOptim automatically handles this gracefully.
16.2 Setup and Imports
- XGBoost (if available)
try:
import xgboost as xgb
XGBOOST_AVAILABLE = True
except ImportError:
XGBOOST_AVAILABLE = False
print("XGBoost not available. Install with: pip install xgboost")
print("Libraries imported successfully!")Libraries imported successfully!16.3 The AWWE Objective Function
We use the Aircraft Wing Weight Example function, which models the weight of an unpainted light aircraft wing. The function accepts inputs in the unit cube \([0,1]^9\) and returns the wing weight in pounds.
from spotoptim.function.so import wingwt
# Problem setup
bounds = [(0, 1)] * 9
param_names = ['Sw', 'Wfw', 'A', 'L', 'q', 'l', 'Rtc', 'Nz', 'Wdg']
max_iter = 20
n_initial = 15
seed = 42
print(f"Problem dimension: {len(bounds)}")
print(f"Optimization budget: {max_iter} evaluations")Problem dimension: 9
Optimization budget: 20 evaluations16.4 1. Default Surrogate: Gaussian Process with Matern Kernel
SpotOptim’s default surrogate is a Gaussian Process with a Matern kernel (ν=2.5), which provides twice-differentiable sample paths and good performance for most optimization problems.
start_time = time.time()
# Default GP (no surrogate specified)
optimizer_default = SpotOptim(
fun=wingwt,
bounds=bounds,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='ei',
seed=seed,
verbose=False
)
result_default = optimizer_default.optimize()
time_default = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_default.fun:.4f} lb")
print(f" Function evaluations: {result_default.nfev}")
print(f" Time: {time_default:.2f}s")
print(f" Success: {result_default.success}")
# Store for comparison
results_comparison = [{
'Surrogate': 'GP Matern ν=2.5 (Default)',
'Best Weight': result_default.fun,
'Evaluations': result_default.nfev,
'Time (s)': time_default,
'Success': result_default.success
}]
Results:
Best weight: 122.1102 lb
Function evaluations: 20
Time: 8.44s
Success: True16.4.1 Visualization: Default Surrogate
# Plot convergence
optimizer_default.plot_progress(log_y=False, figsize=(10, 5))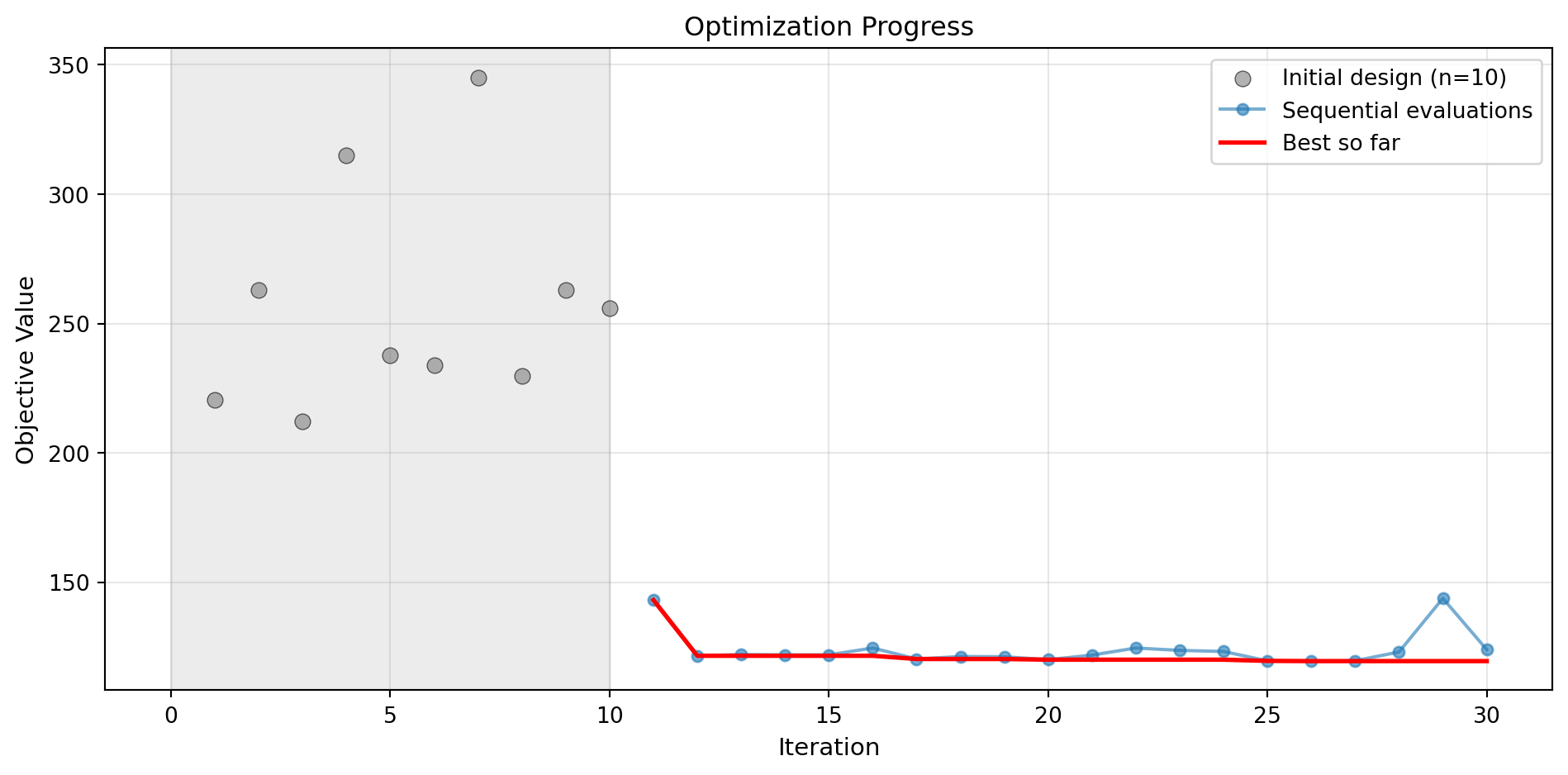
# Plot most important hyperparameters
optimizer_default.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('Default GP Matern ν=2.5: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Sw: importance = 19.81% (type: float)
Nz: importance = 18.67% (type: float)
Rtc: importance = 17.96% (type: float)
Generating 3 surrogate plots...
Plotting Sw vs Nz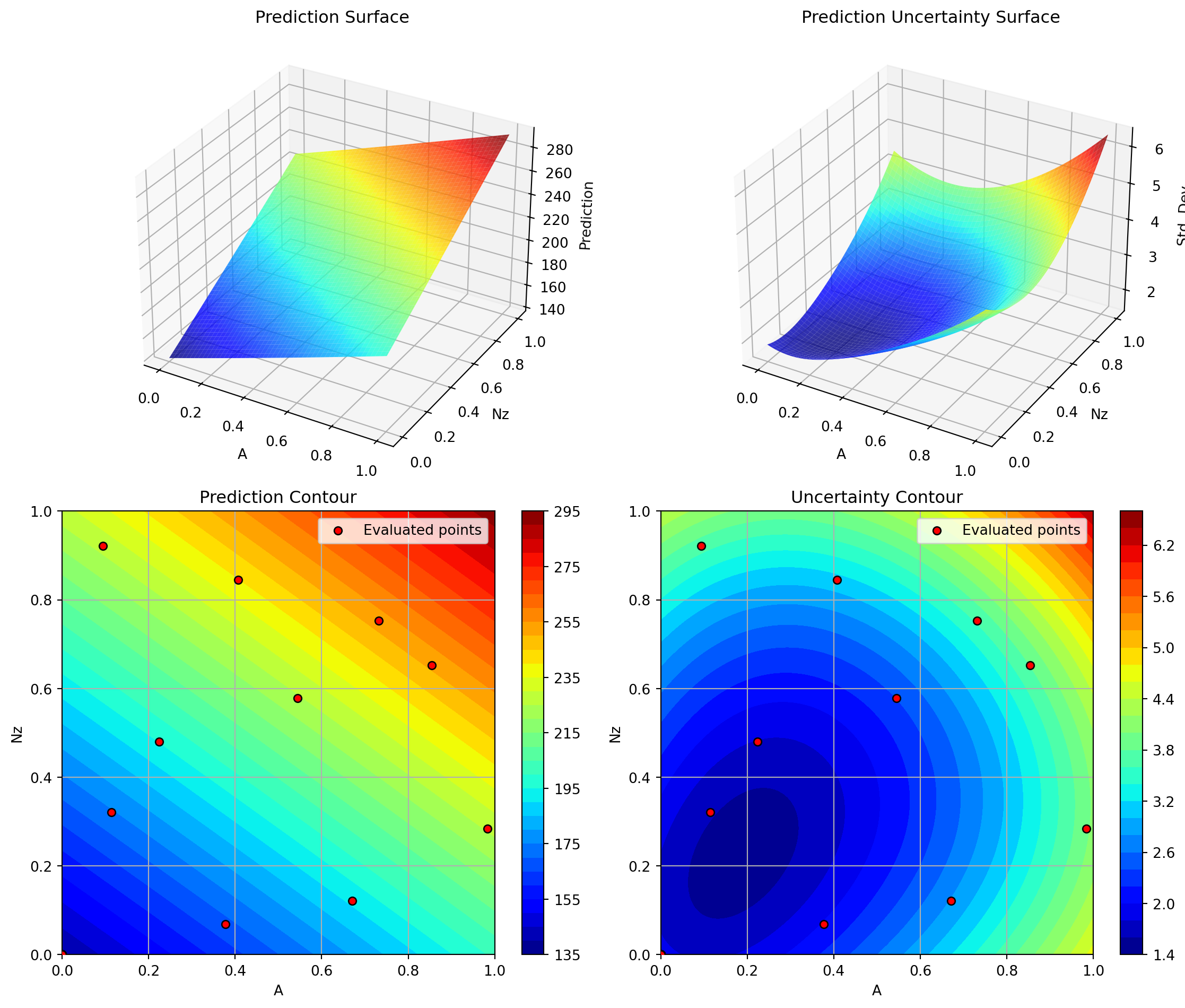
Plotting Sw vs Rtc
Plotting Nz vs Rtc
<Figure size 672x480 with 0 Axes>16.5 2. Gaussian Process with RBF (Radial Basis Function) Kernel
The RBF kernel (also called squared exponential) produces infinitely differentiable sample paths, resulting in very smooth predictions.
start_time = time.time()
# Configure GP with RBF kernel
kernel_rbf = ConstantKernel(1.0, (1e-3, 1e3)) * RBF(
length_scale=1.0,
length_scale_bounds=(1e-2, 1e2)
)
gp_rbf = GaussianProcessRegressor(
kernel=kernel_rbf,
n_restarts_optimizer=10,
normalize_y=True,
random_state=seed
)
optimizer_rbf = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=gp_rbf,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='ei',
seed=seed,
verbose=False
)
result_rbf = optimizer_rbf.optimize()
time_rbf = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_rbf.fun:.4f} lb")
print(f" Function evaluations: {result_rbf.nfev}")
print(f" Time: {time_rbf:.2f}s")
print(f" Success: {result_rbf.success}")
results_comparison.append({
'Surrogate': 'GP RBF',
'Best Weight': result_rbf.fun,
'Evaluations': result_rbf.nfev,
'Time (s)': time_rbf,
'Success': result_rbf.success
})
Results:
Best weight: 121.3024 lb
Function evaluations: 20
Time: 7.38s
Success: True16.5.1 Visualization: RBF Kernel
optimizer_rbf.plot_progress(log_y=False, figsize=(10, 5))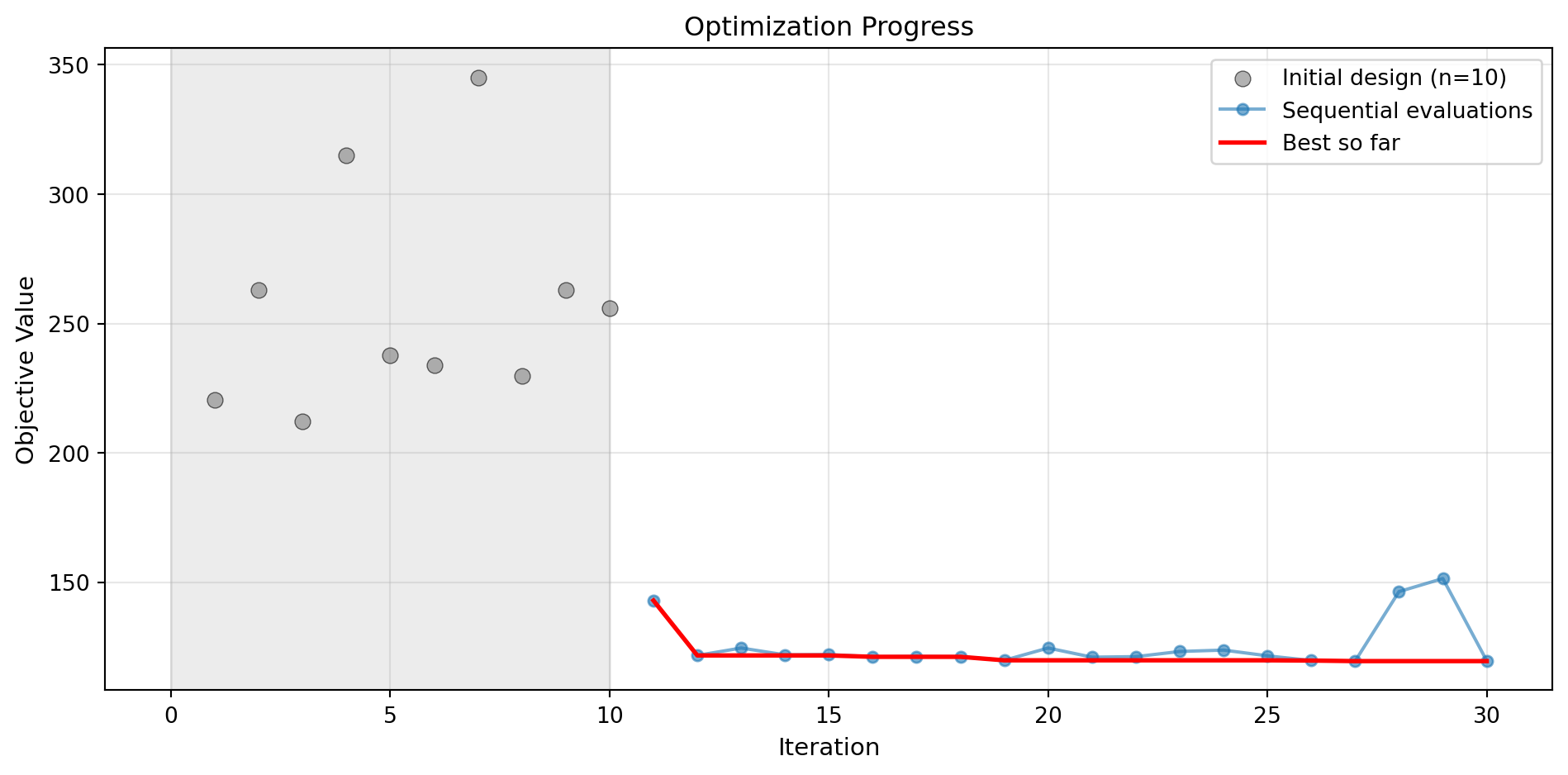
optimizer_rbf.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('GP RBF Kernel: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Sw: importance = 18.38% (type: float)
Nz: importance = 17.32% (type: float)
Rtc: importance = 16.67% (type: float)
Generating 3 surrogate plots...
Plotting Sw vs Nz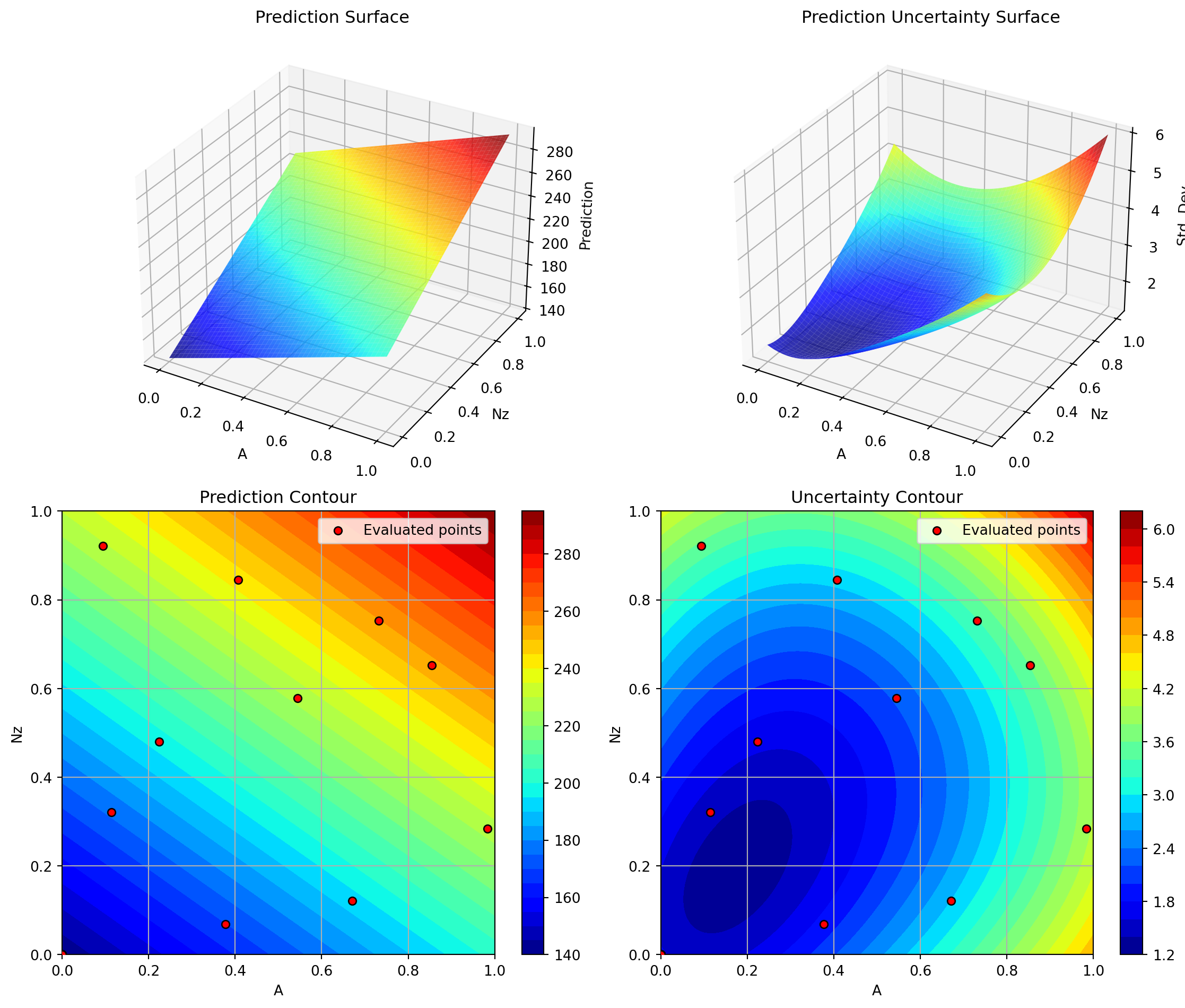
Plotting Sw vs Rtc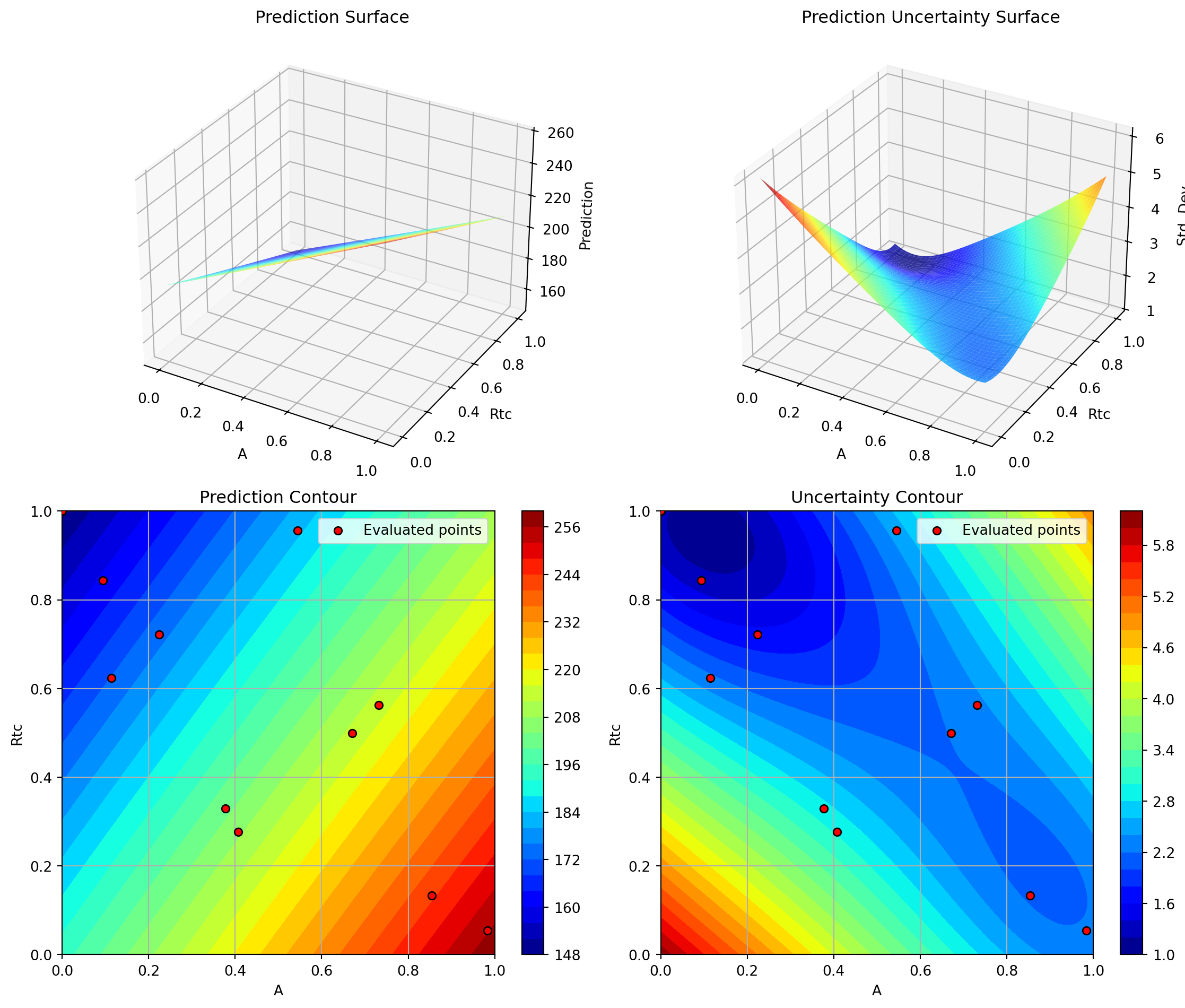
Plotting Nz vs Rtc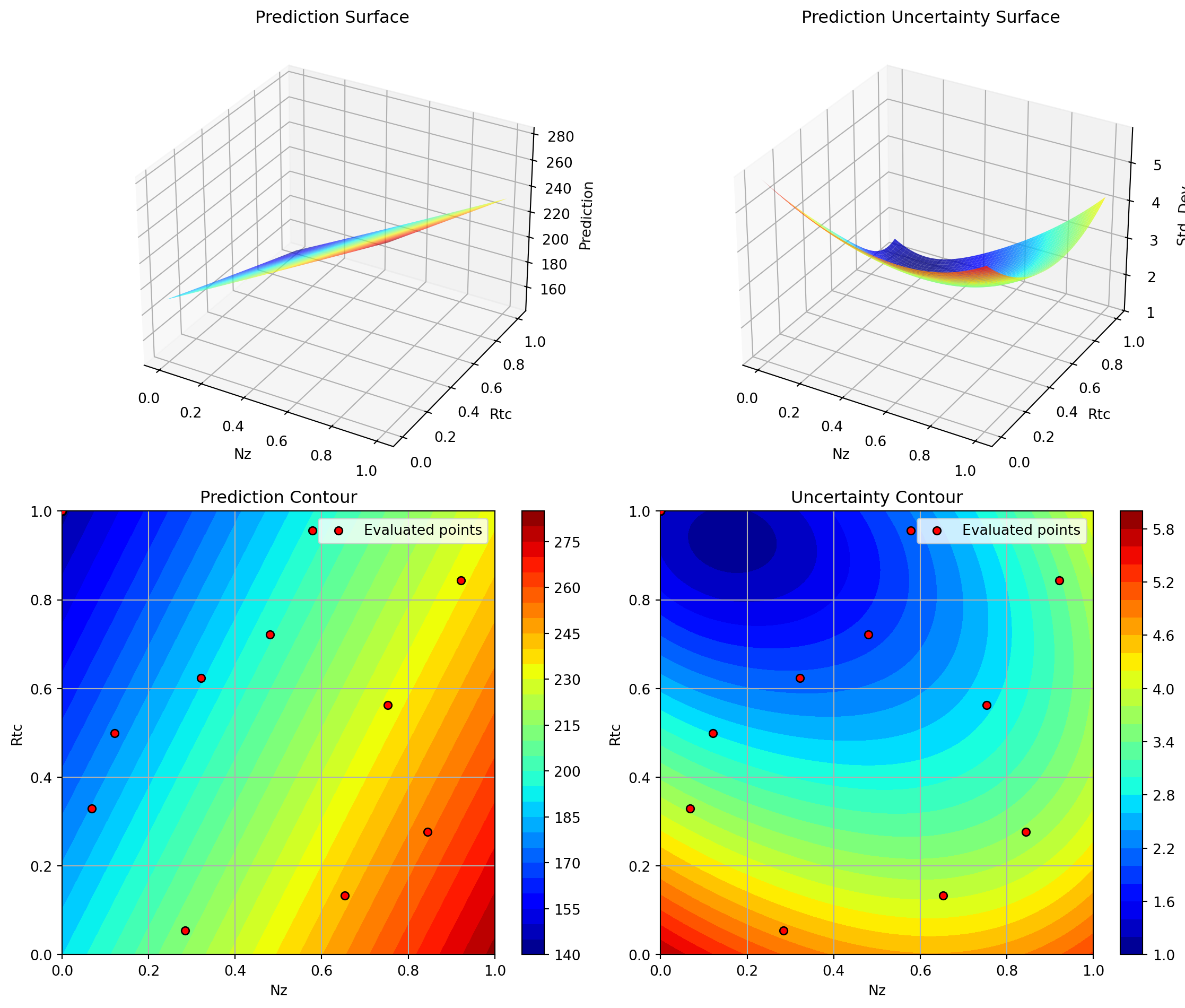
<Figure size 672x480 with 0 Axes>16.6 3. Gaussian Process with Matern ν=1.5 Kernel
The Matern ν=1.5 kernel produces once-differentiable sample paths, allowing for more flexible (less smooth) fits than ν=2.5.
start_time = time.time()
# Configure GP with Matern nu=1.5
kernel_matern15 = ConstantKernel(1.0, (1e-3, 1e3)) * Matern(
length_scale=1.0,
length_scale_bounds=(1e-2, 1e2),
nu=1.5
)
gp_matern15 = GaussianProcessRegressor(
kernel=kernel_matern15,
n_restarts_optimizer=10,
normalize_y=True,
random_state=seed
)
optimizer_matern15 = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=gp_matern15,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='ei',
seed=seed,
verbose=False
)
result_matern15 = optimizer_matern15.optimize()
time_matern15 = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_matern15.fun:.4f} lb")
print(f" Function evaluations: {result_matern15.nfev}")
print(f" Time: {time_matern15:.2f}s")
print(f" Success: {result_matern15.success}")
results_comparison.append({
'Surrogate': 'GP Matern ν=1.5',
'Best Weight': result_matern15.fun,
'Evaluations': result_matern15.nfev,
'Time (s)': time_matern15,
'Success': result_matern15.success
})
Results:
Best weight: 121.2565 lb
Function evaluations: 20
Time: 6.88s
Success: True16.6.1 Visualization: Matern ν=1.5 Kernel
optimizer_matern15.plot_progress(log_y=False, figsize=(10, 5))
optimizer_matern15.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('GP Matern ν=1.5 Kernel: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Sw: importance = 19.76% (type: float)
Nz: importance = 18.63% (type: float)
Rtc: importance = 17.93% (type: float)
Generating 3 surrogate plots...
Plotting Sw vs Nz
Plotting Sw vs Rtc
Plotting Nz vs Rtc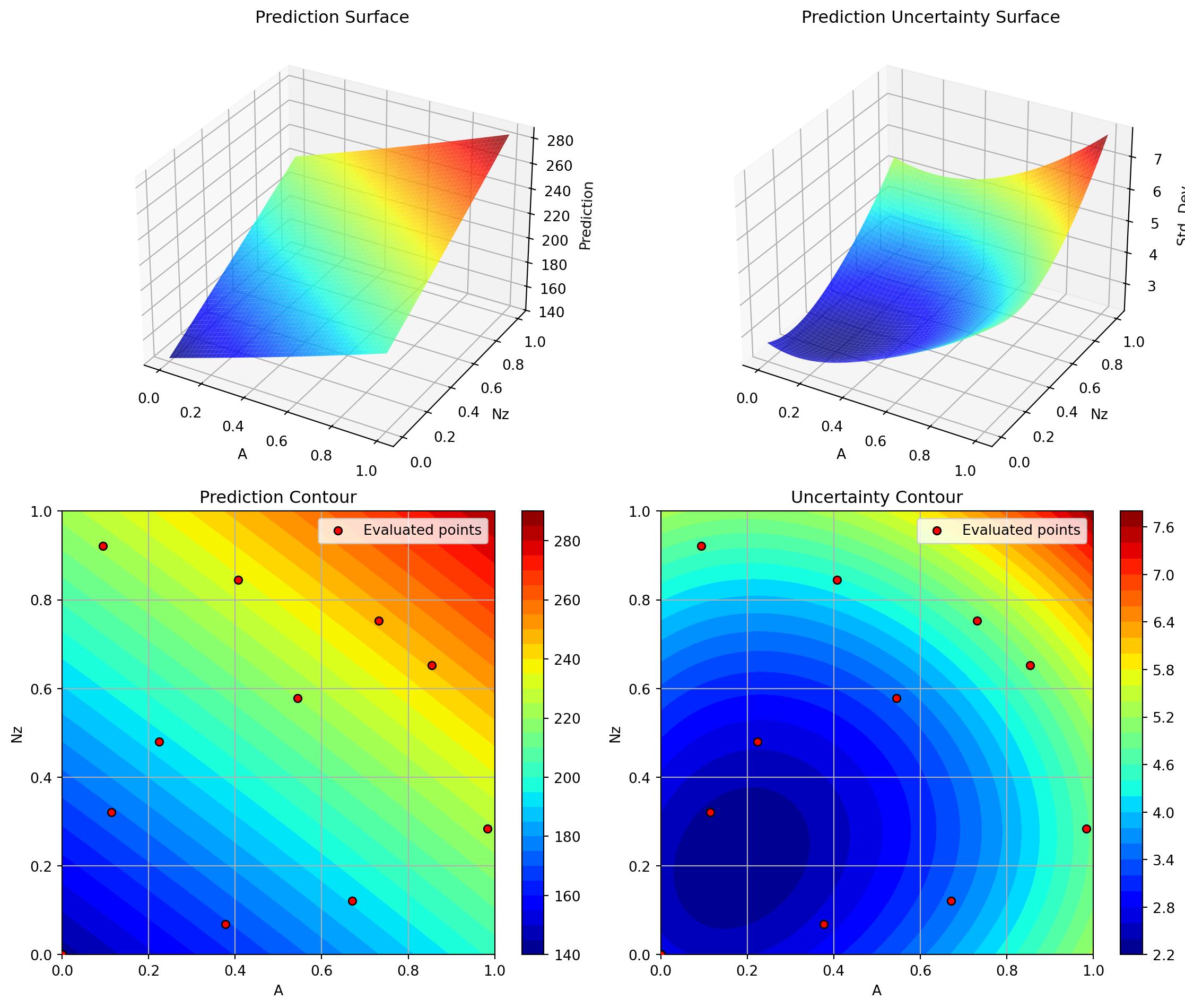
<Figure size 672x480 with 0 Axes>16.7 4. Gaussian Process with Rational Quadratic Kernel
The Rational Quadratic kernel is a scale mixture of RBF kernels with different length scales, providing more flexibility than a single RBF.
start_time = time.time()
# Configure GP with Rational Quadratic kernel
kernel_rq = ConstantKernel(1.0, (1e-3, 1e3)) * RationalQuadratic(
length_scale=1.0,
alpha=1.0,
length_scale_bounds=(1e-2, 1e2),
alpha_bounds=(1e-2, 1e2)
)
gp_rq = GaussianProcessRegressor(
kernel=kernel_rq,
n_restarts_optimizer=10,
normalize_y=True,
random_state=seed
)
optimizer_rq = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=gp_rq,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='ei',
seed=seed,
verbose=False
)
result_rq = optimizer_rq.optimize()
time_rq = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_rq.fun:.4f} lb")
print(f" Function evaluations: {result_rq.nfev}")
print(f" Time: {time_rq:.2f}s")
print(f" Success: {result_rq.success}")
results_comparison.append({
'Surrogate': 'GP Rational Quadratic',
'Best Weight': result_rq.fun,
'Evaluations': result_rq.nfev,
'Time (s)': time_rq,
'Success': result_rq.success
})
Results:
Best weight: 121.2592 lb
Function evaluations: 20
Time: 6.45s
Success: True16.7.1 Visualization: Rational Quadratic Kernel
optimizer_rq.plot_progress(log_y=False, figsize=(10, 5))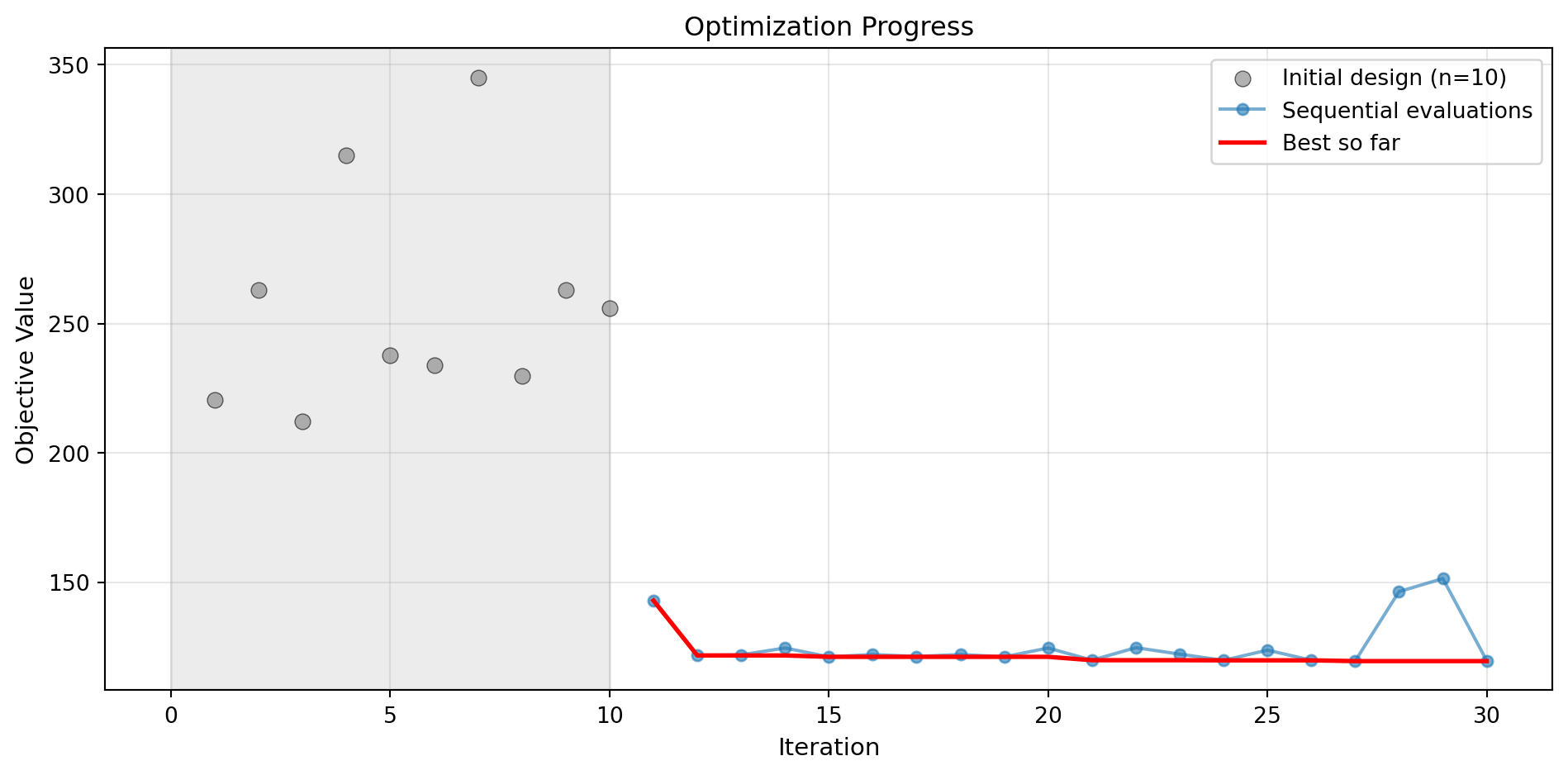
optimizer_rq.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('GP Rational Quadratic Kernel: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Sw: importance = 17.61% (type: float)
Nz: importance = 16.60% (type: float)
Rtc: importance = 15.98% (type: float)
Generating 3 surrogate plots...
Plotting Sw vs Nz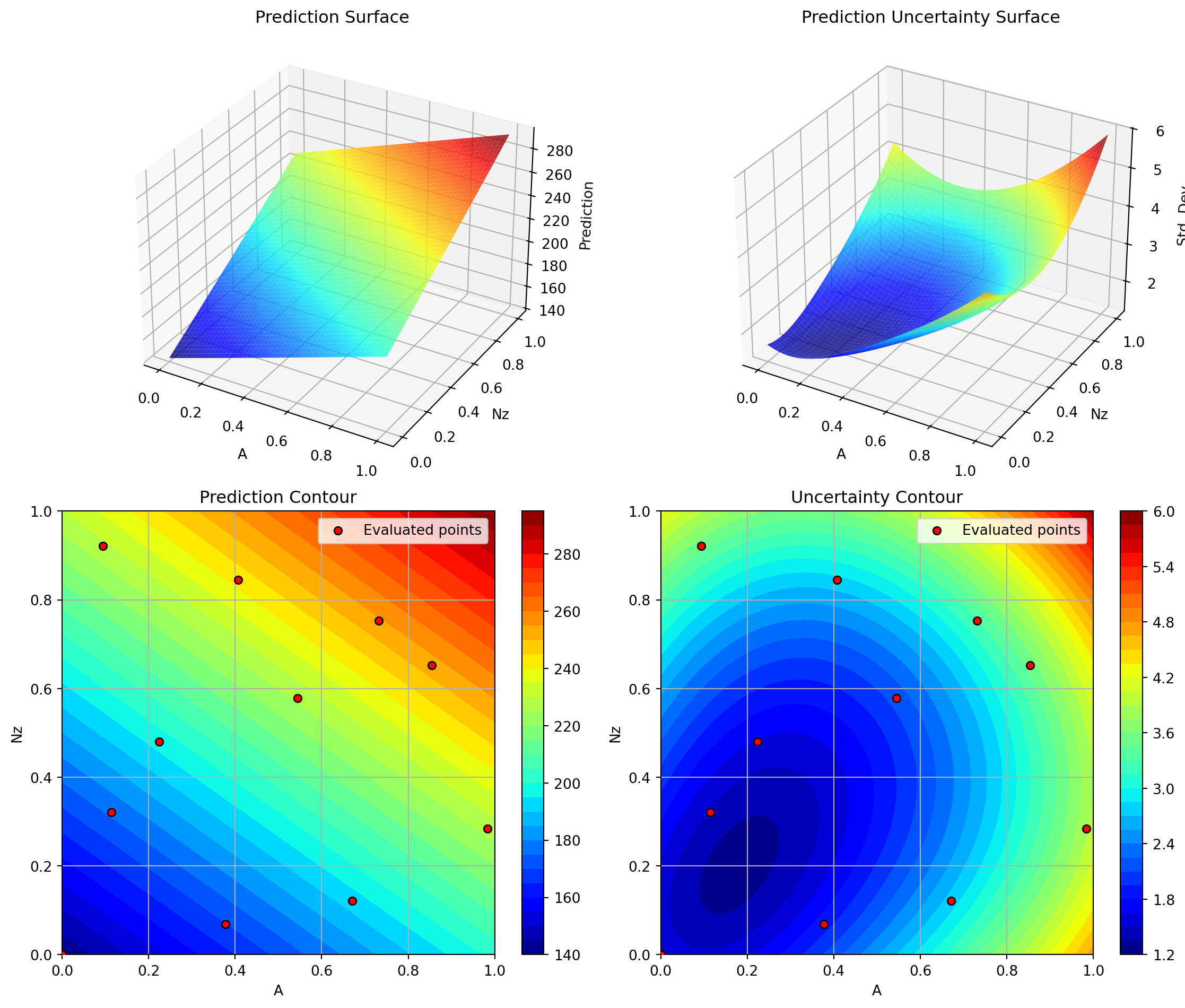
Plotting Sw vs Rtc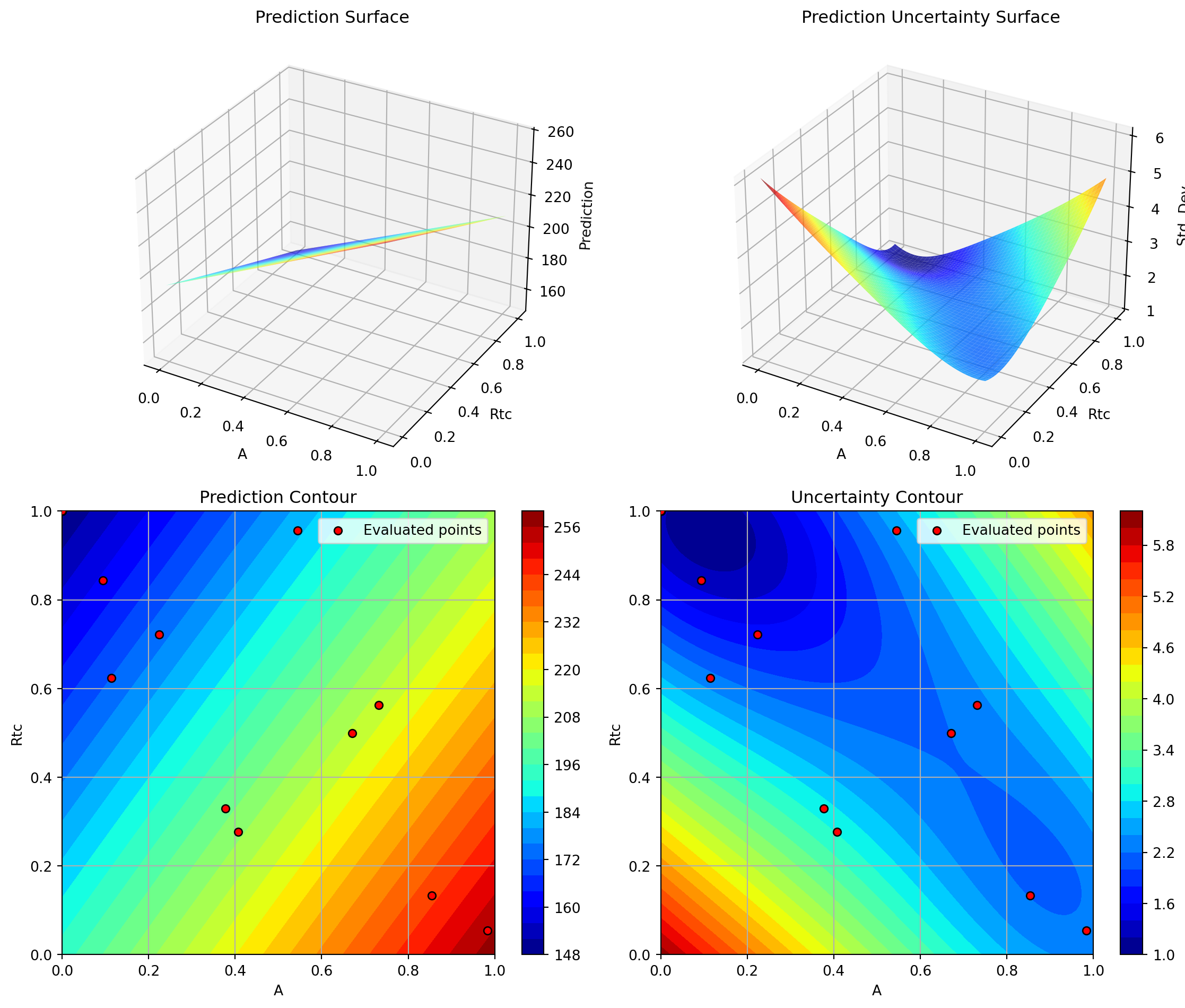
Plotting Nz vs Rtc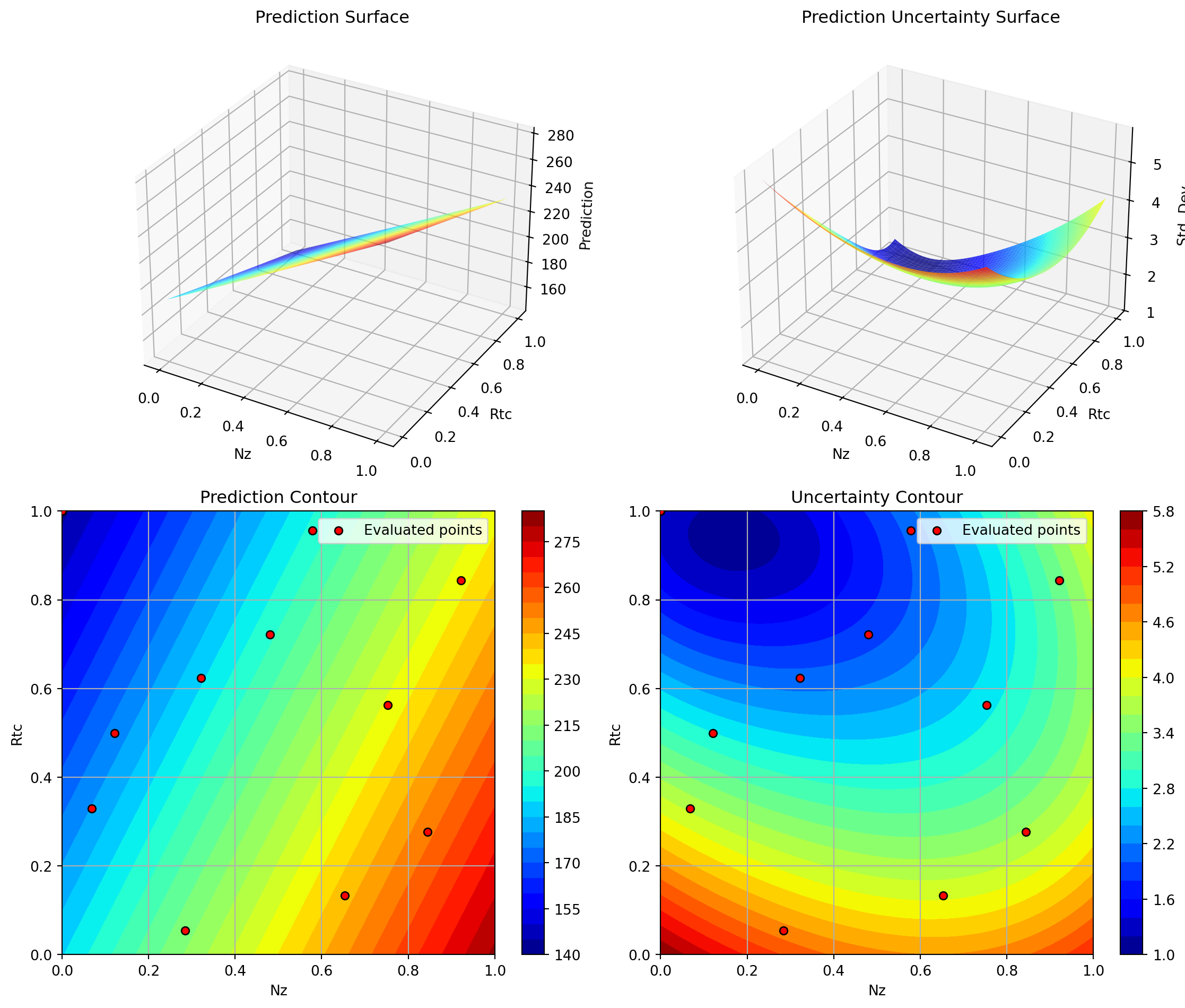
<Figure size 672x480 with 0 Axes>16.8 5. SpotOptim Kriging Model
SpotOptim includes its own Kriging implementation optimized for sequential design. It uses Gaussian correlation function and optimizes hyperparameters via differential evolution.
start_time = time.time()
# Configure Kriging model
kriging_model = Kriging(
noise=1e-10, # Regularization parameter
kernel='gauss', # Gaussian/RBF kernel
n_theta=None, # Auto: use number of dimensions
min_theta=-3.0, # Min log10(theta) bound
max_theta=2.0, # Max log10(theta) bound
seed=seed
)
optimizer_kriging = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=kriging_model,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='ei',
seed=seed,
verbose=False
)
result_kriging = optimizer_kriging.optimize()
time_kriging = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_kriging.fun:.4f} lb")
print(f" Function evaluations: {result_kriging.nfev}")
print(f" Time: {time_kriging:.2f}s")
print(f" Success: {result_kriging.success}")
results_comparison.append({
'Surrogate': 'SpotOptim Kriging',
'Best Weight': result_kriging.fun,
'Evaluations': result_kriging.nfev,
'Time (s)': time_kriging,
'Success': result_kriging.success
})
Results:
Best weight: 121.1616 lb
Function evaluations: 20
Time: 13.56s
Success: True16.8.1 Visualization: Kriging Model
optimizer_kriging.plot_progress(log_y=False, figsize=(10, 5))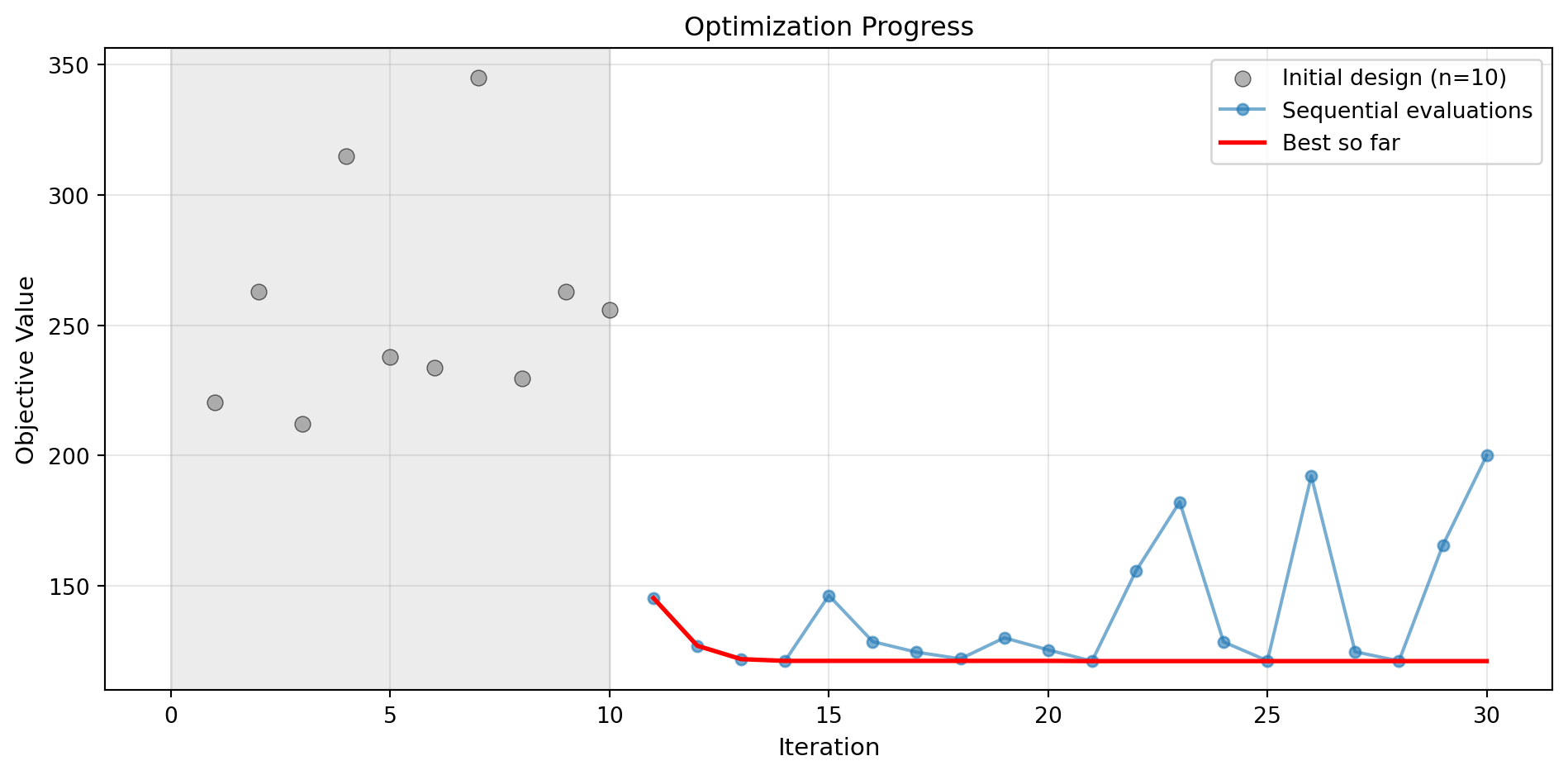
optimizer_kriging.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('SpotOptim Kriging: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Sw: importance = 14.87% (type: float)
Nz: importance = 14.01% (type: float)
A: importance = 13.44% (type: float)
Generating 3 surrogate plots...
Plotting Sw vs Nz
Plotting Sw vs A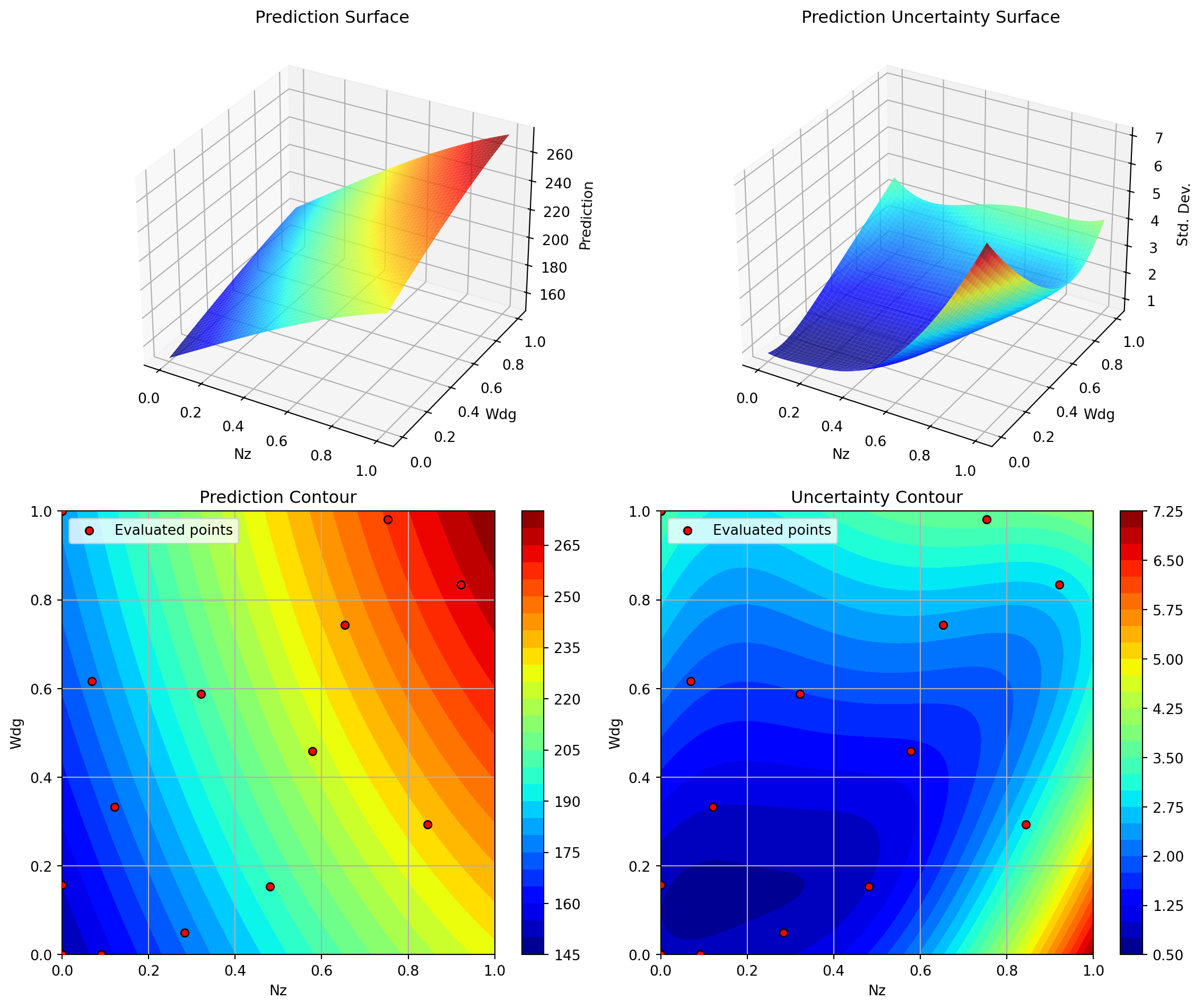
Plotting Nz vs A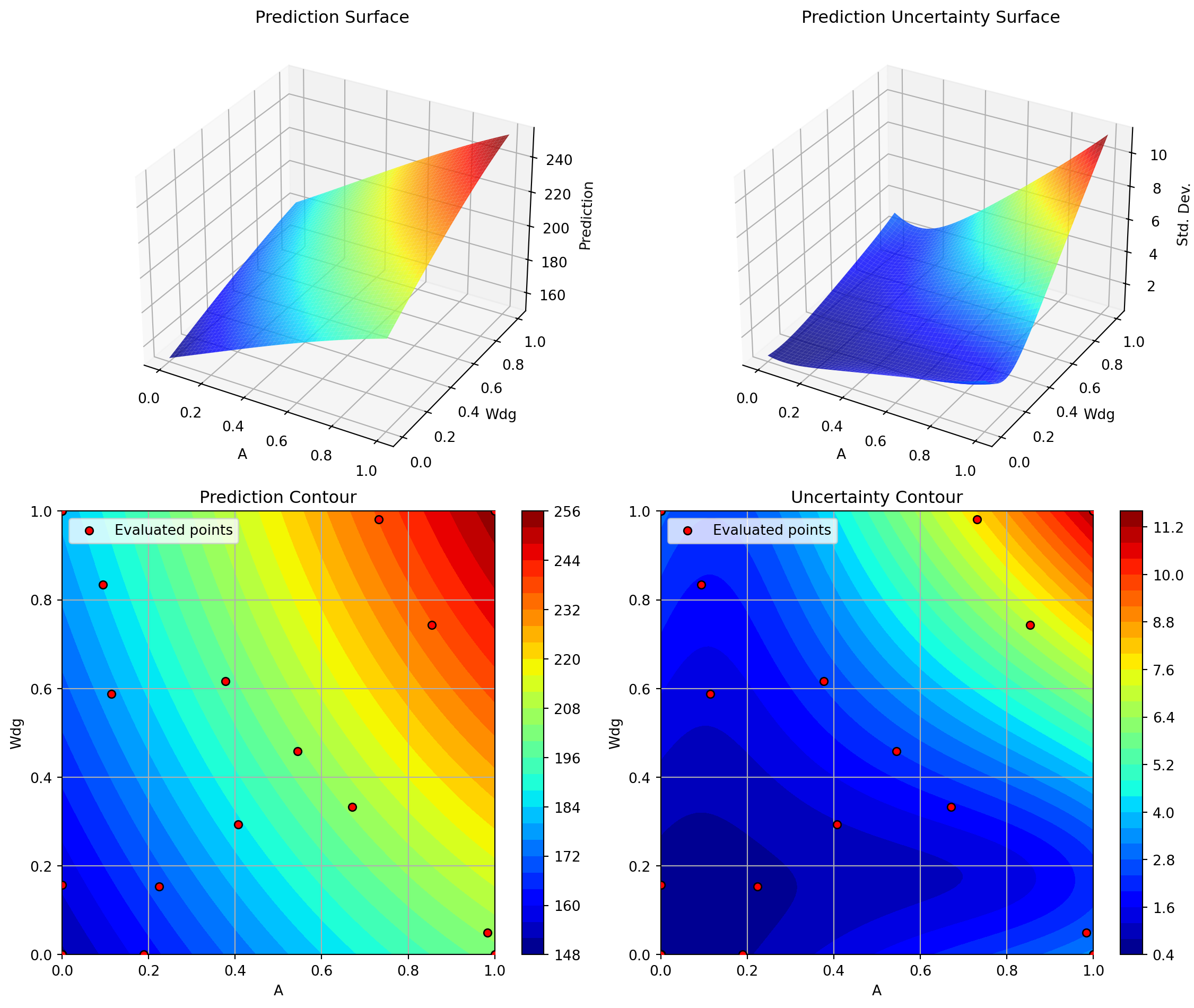
<Figure size 672x480 with 0 Axes>16.9 6. Random Forest Regressor
Random Forests are ensemble methods that handle noise well and can model discontinuities. They don’t naturally provide uncertainty estimates, so the acquisition function uses predictions only.
start_time = time.time()
# Configure Random Forest
rf_model = RandomForestRegressor(
n_estimators=100,
max_depth=15,
min_samples_split=2,
min_samples_leaf=1,
random_state=seed,
)
optimizer_rf = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=rf_model,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='y', # Use 'y' (greedy) since RF doesn't provide std
seed=seed,
verbose=False
)
result_rf = optimizer_rf.optimize()
time_rf = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_rf.fun:.4f} lb")
print(f" Function evaluations: {result_rf.nfev}")
print(f" Time: {time_rf:.2f}s")
print(f" Success: {result_rf.success}")
print(f" Note: Using acquisition='y' (greedy) since RF doesn't provide uncertainty")
results_comparison.append({
'Surrogate': 'Random Forest',
'Best Weight': result_rf.fun,
'Evaluations': result_rf.nfev,
'Time (s)': time_rf,
'Success': result_rf.success
})
Results:
Best weight: 150.4965 lb
Function evaluations: 20
Time: 57.37s
Success: True
Note: Using acquisition='y' (greedy) since RF doesn't provide uncertainty16.9.1 Visualization: Random Forest
optimizer_rf.plot_progress(log_y=False, figsize=(10, 5))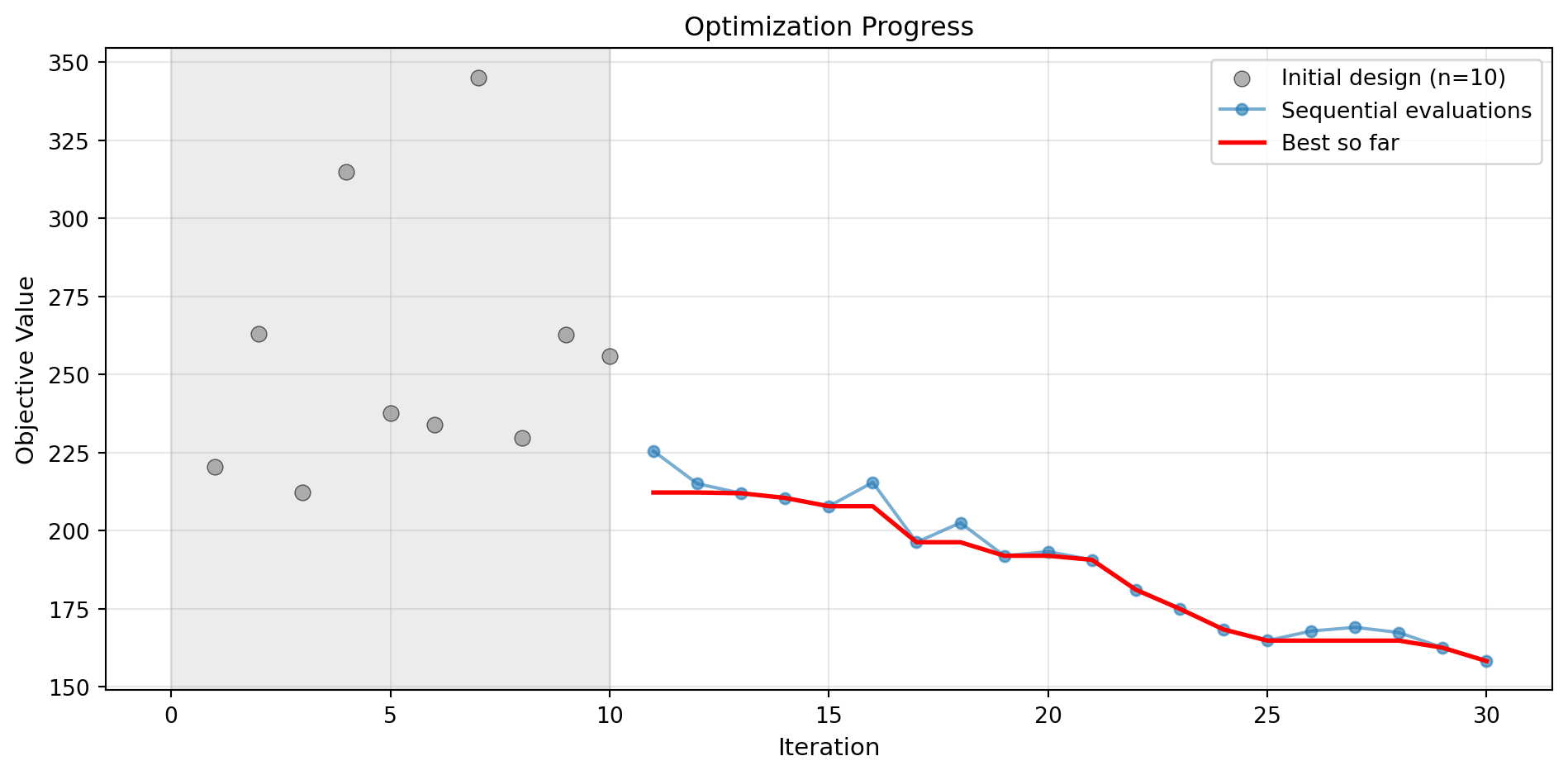
optimizer_rf.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('Random Forest: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Nz: importance = 15.09% (type: float)
A: importance = 14.50% (type: float)
Wfw: importance = 12.13% (type: float)
Generating 3 surrogate plots...
Plotting Nz vs A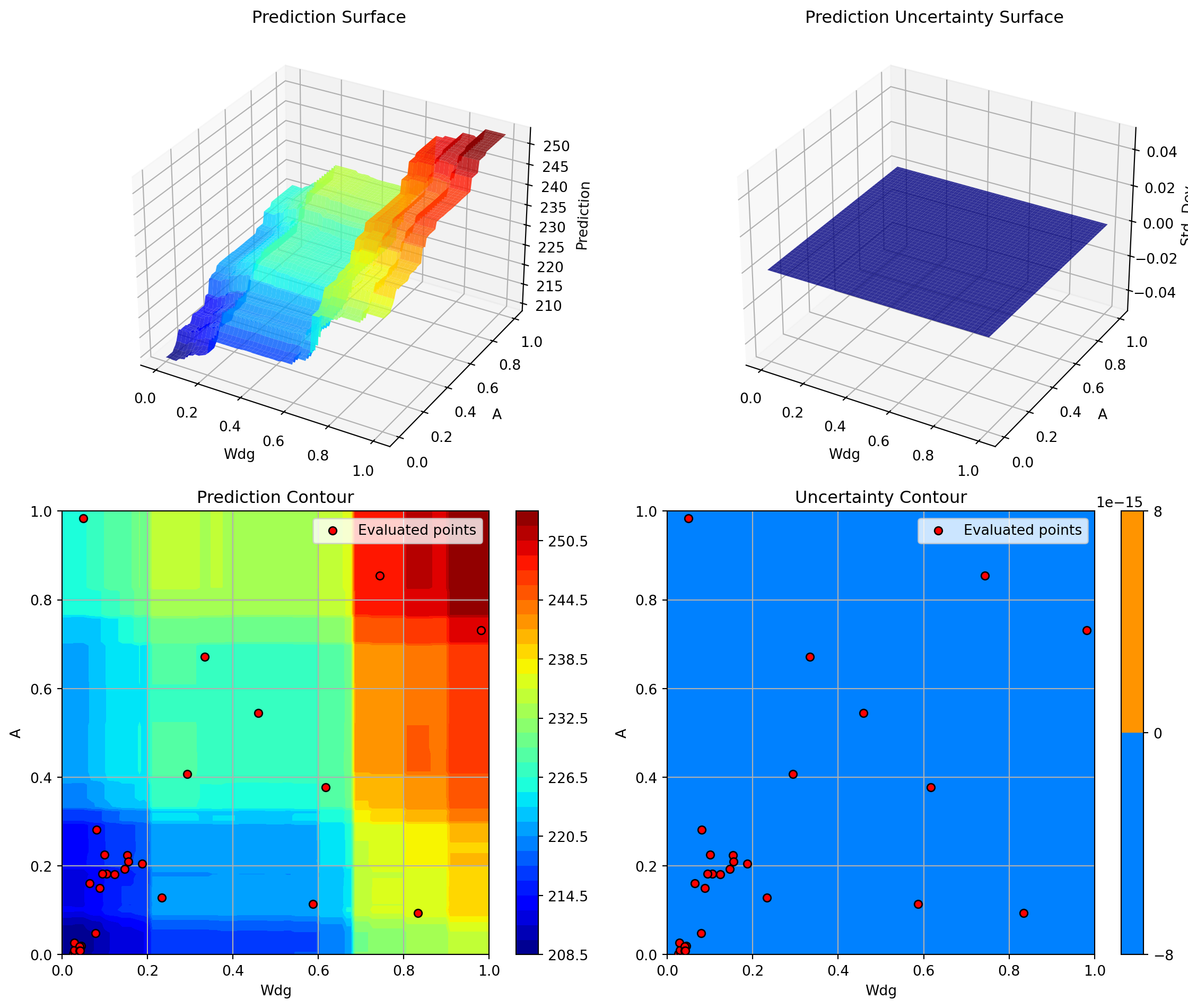
Plotting Nz vs Wfw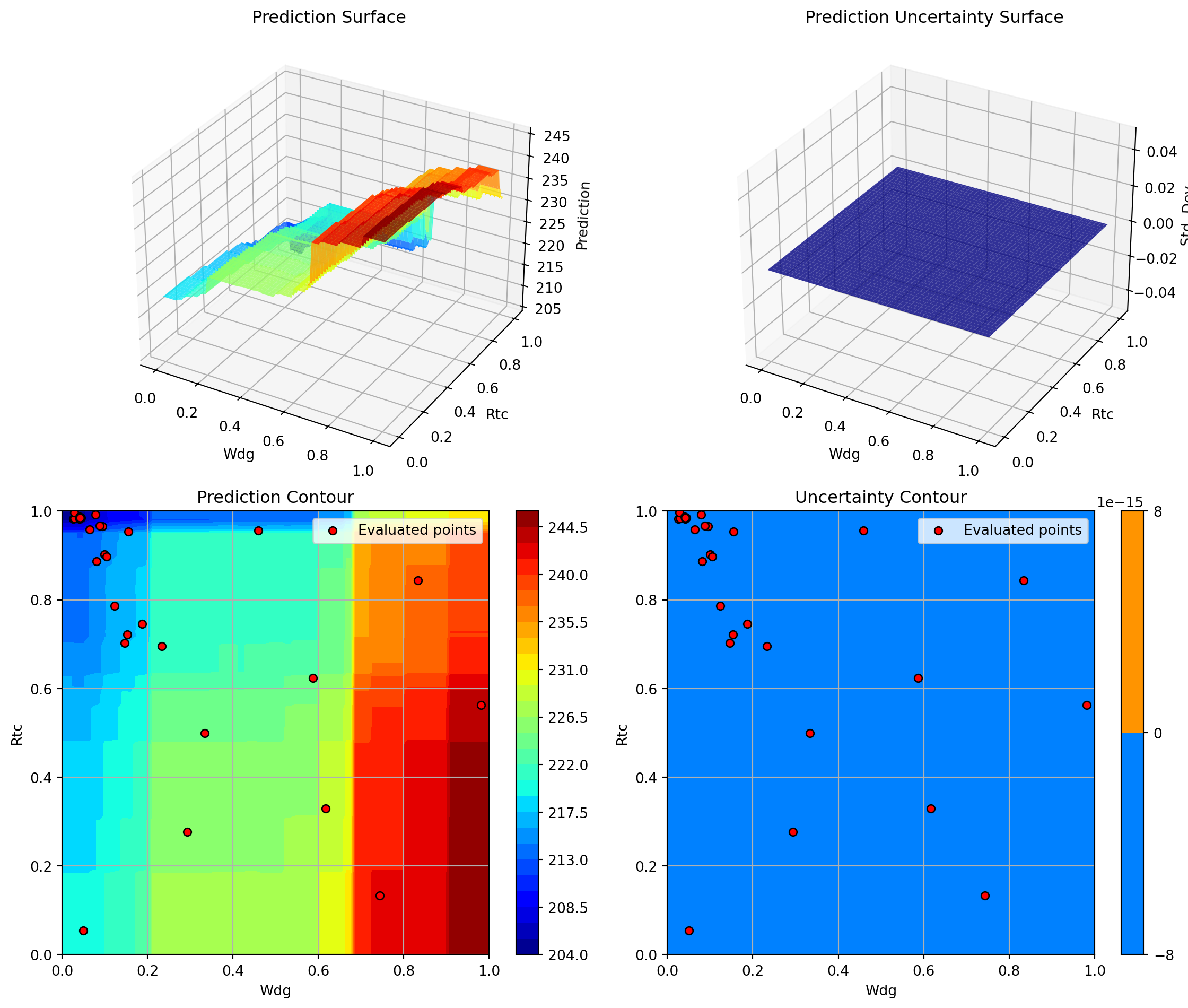
Plotting A vs Wfw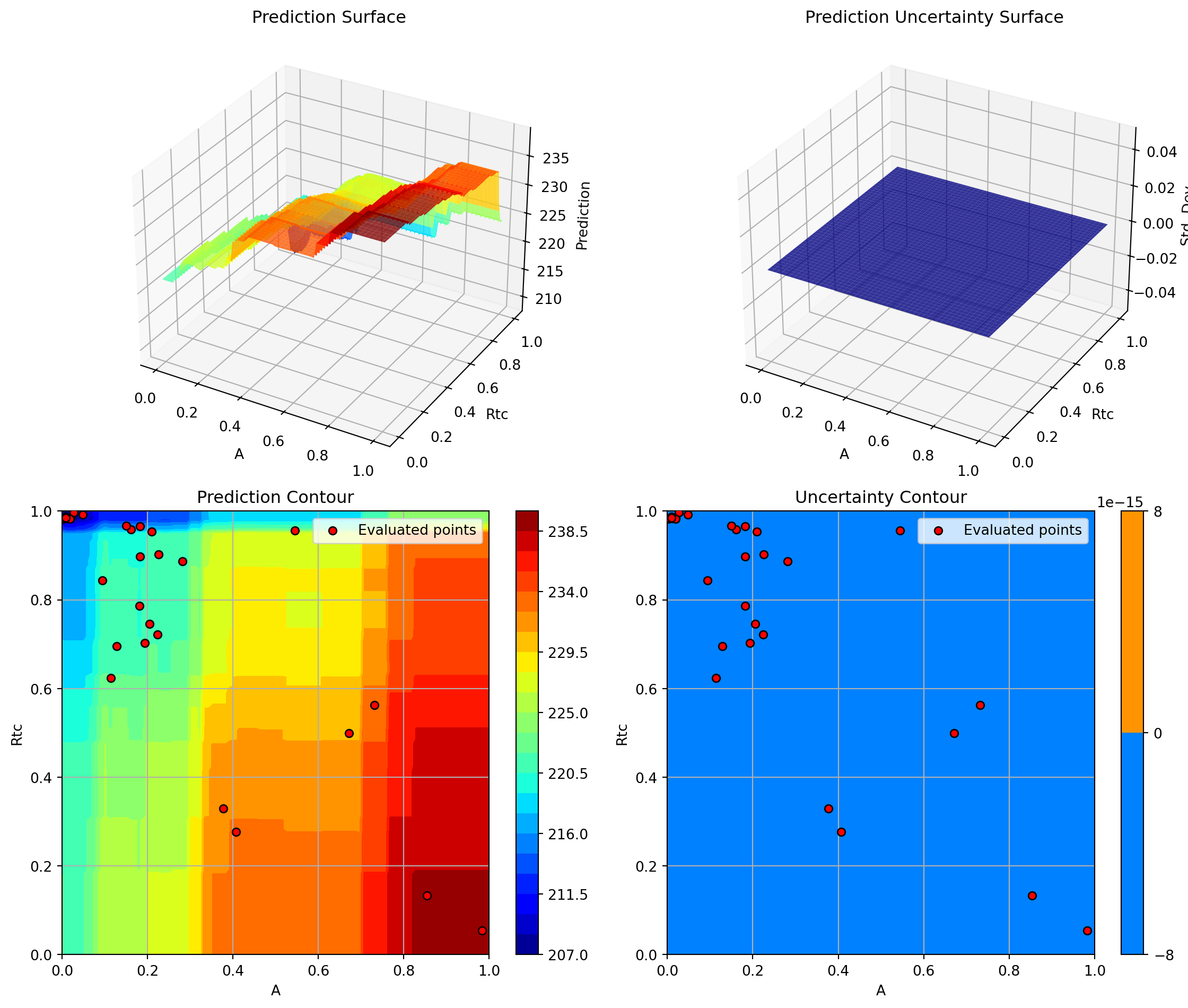
<Figure size 672x480 with 0 Axes>16.10 7. XGBoost Regressor
XGBoost is a gradient boosting implementation known for excellent performance on structured data and fast training/prediction times.
if XGBOOST_AVAILABLE:
start_time = time.time()
# Configure XGBoost
xgb_model = xgb.XGBRegressor(
n_estimators=100,
max_depth=6,
learning_rate=0.1,
subsample=0.8,
colsample_bytree=0.8,
random_state=seed,
)
optimizer_xgb = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=xgb_model,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='y', # Use 'y' (greedy) since XGBoost doesn't provide std
seed=seed,
verbose=False
)
result_xgb = optimizer_xgb.optimize()
time_xgb = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_xgb.fun:.4f} lb")
print(f" Function evaluations: {result_xgb.nfev}")
print(f" Time: {time_xgb:.2f}s")
print(f" Success: {result_xgb.success}")
print(f" Note: Using acquisition='y' (greedy) since XGBoost doesn't provide uncertainty")
results_comparison.append({
'Surrogate': 'XGBoost',
'Best Weight': result_xgb.fun,
'Evaluations': result_xgb.nfev,
'Time (s)': time_xgb,
'Success': result_xgb.success
})
# Visualization
optimizer_xgb.plot_progress(log_y=False, figsize=(10, 5))
plt.title('XGBoost: Convergence')
plt.show()
optimizer_xgb.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('XGBoost: Most Important Parameters', y=1.02)
plt.show()
else:
print("=" * 80)
print("7. XGBoost Regressor - SKIPPED (not installed)")
print("=" * 80)
print("Install XGBoost with: pip install xgboost")
Results:
Best weight: 143.0885 lb
Function evaluations: 20
Time: 5.09s
Success: True
Note: Using acquisition='y' (greedy) since XGBoost doesn't provide uncertainty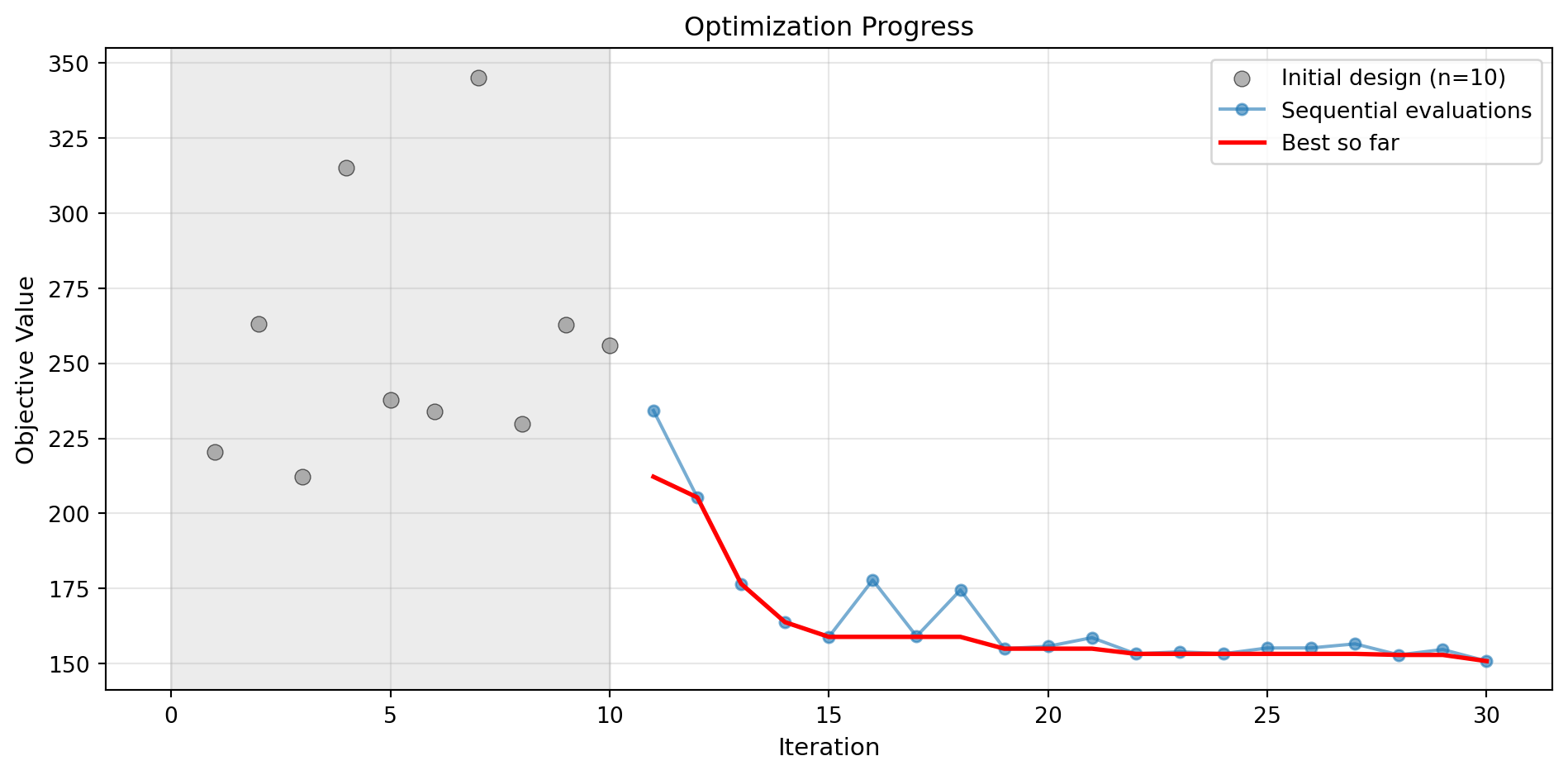
Plotting surrogate contours for top 3 most important parameters:
Nz: importance = 17.11% (type: float)
Sw: importance = 14.26% (type: float)
Rtc: importance = 14.00% (type: float)
Generating 3 surrogate plots...
Plotting Nz vs Sw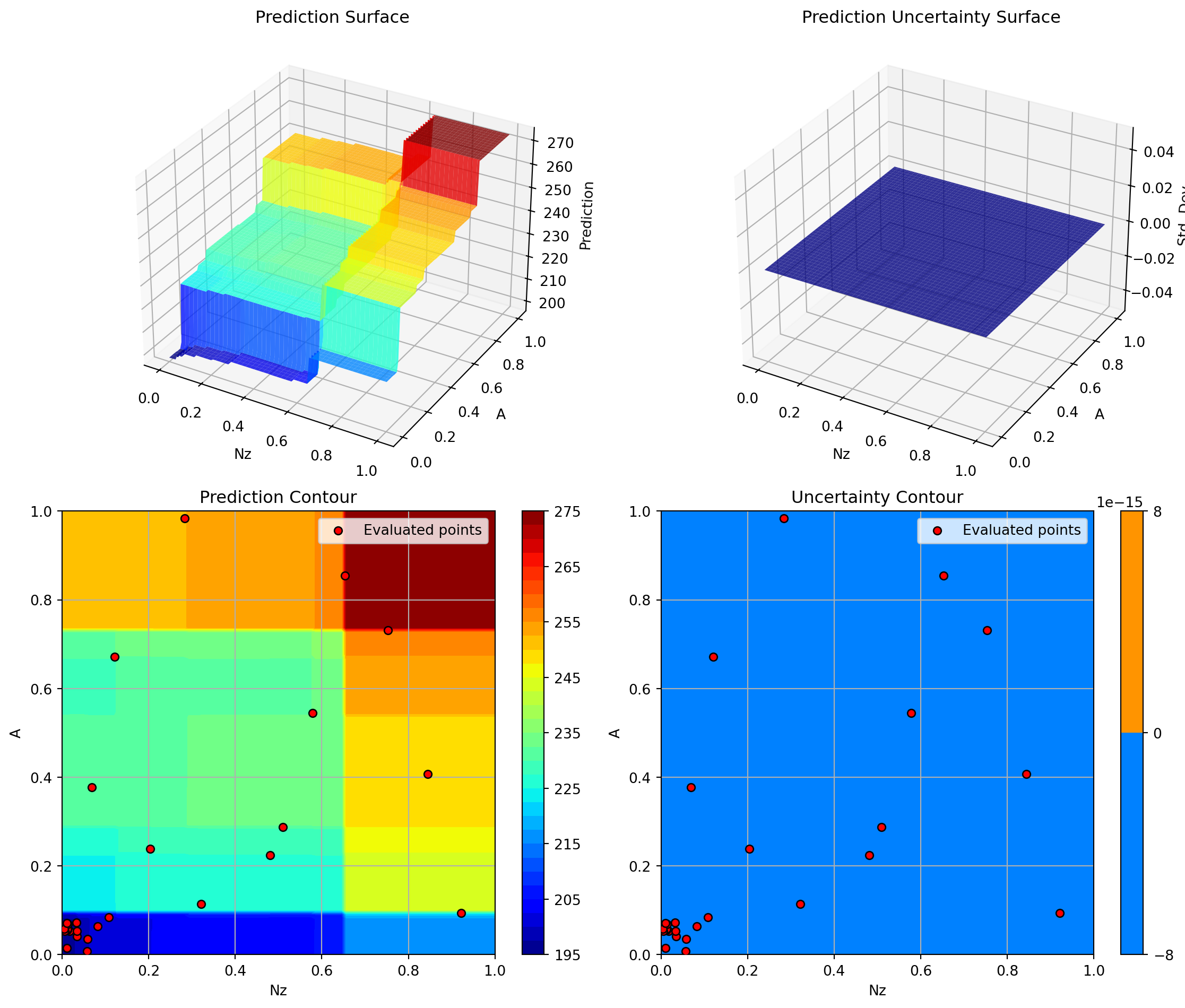
Plotting Nz vs Rtc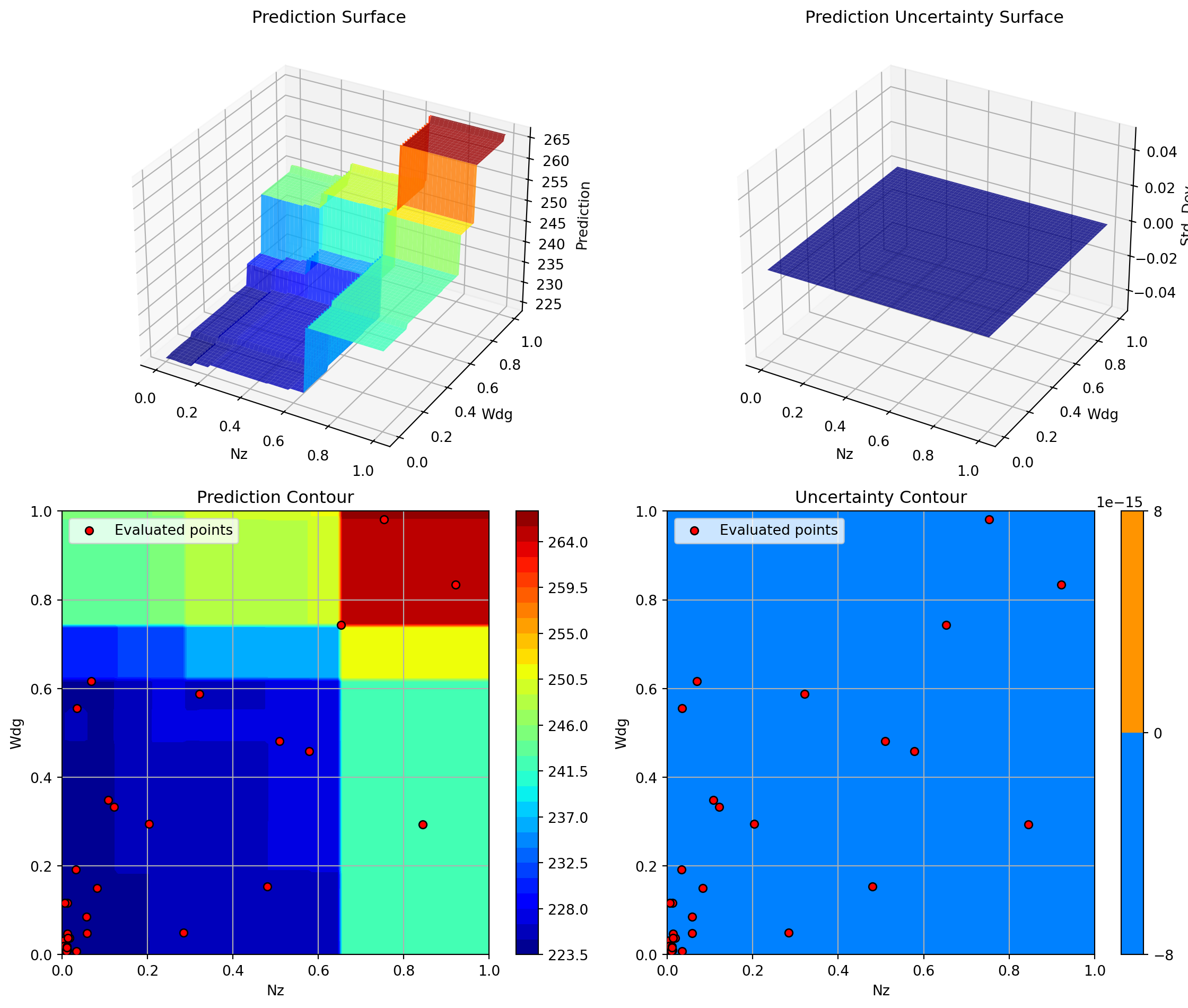
Plotting Sw vs Rtc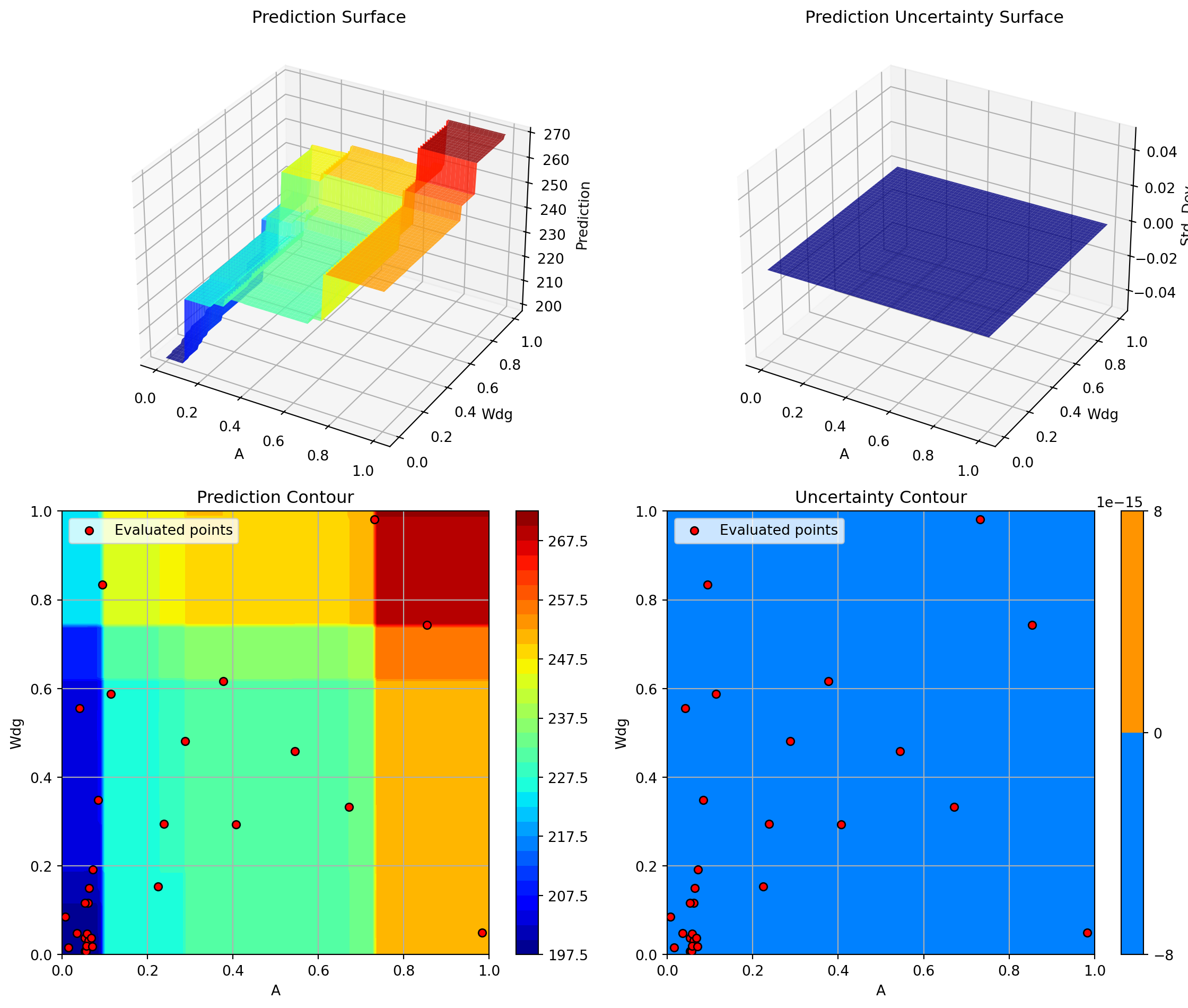
<Figure size 672x480 with 0 Axes>16.11 8. Support Vector Regression (SVR)
SVR with RBF kernel can model complex non-linear relationships. It’s particularly good for high-dimensional problems with smooth structure.
start_time = time.time()
# Configure SVR
svr_model = SVR(
kernel='rbf',
C=100.0,
epsilon=0.1,
gamma='scale'
)
optimizer_svr = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=svr_model,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='y', # Use 'y' (greedy) since SVR doesn't provide std by default
seed=seed,
verbose=False
)
result_svr = optimizer_svr.optimize()
time_svr = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_svr.fun:.4f} lb")
print(f" Function evaluations: {result_svr.nfev}")
print(f" Time: {time_svr:.2f}s")
print(f" Success: {result_svr.success}")
results_comparison.append({
'Surrogate': 'SVR (RBF)',
'Best Weight': result_svr.fun,
'Evaluations': result_svr.nfev,
'Time (s)': time_svr,
'Success': result_svr.success
})
Results:
Best weight: 156.8869 lb
Function evaluations: 20
Time: 0.52s
Success: True16.11.1 Visualization: SVR
optimizer_svr.plot_progress(log_y=False, figsize=(10, 5))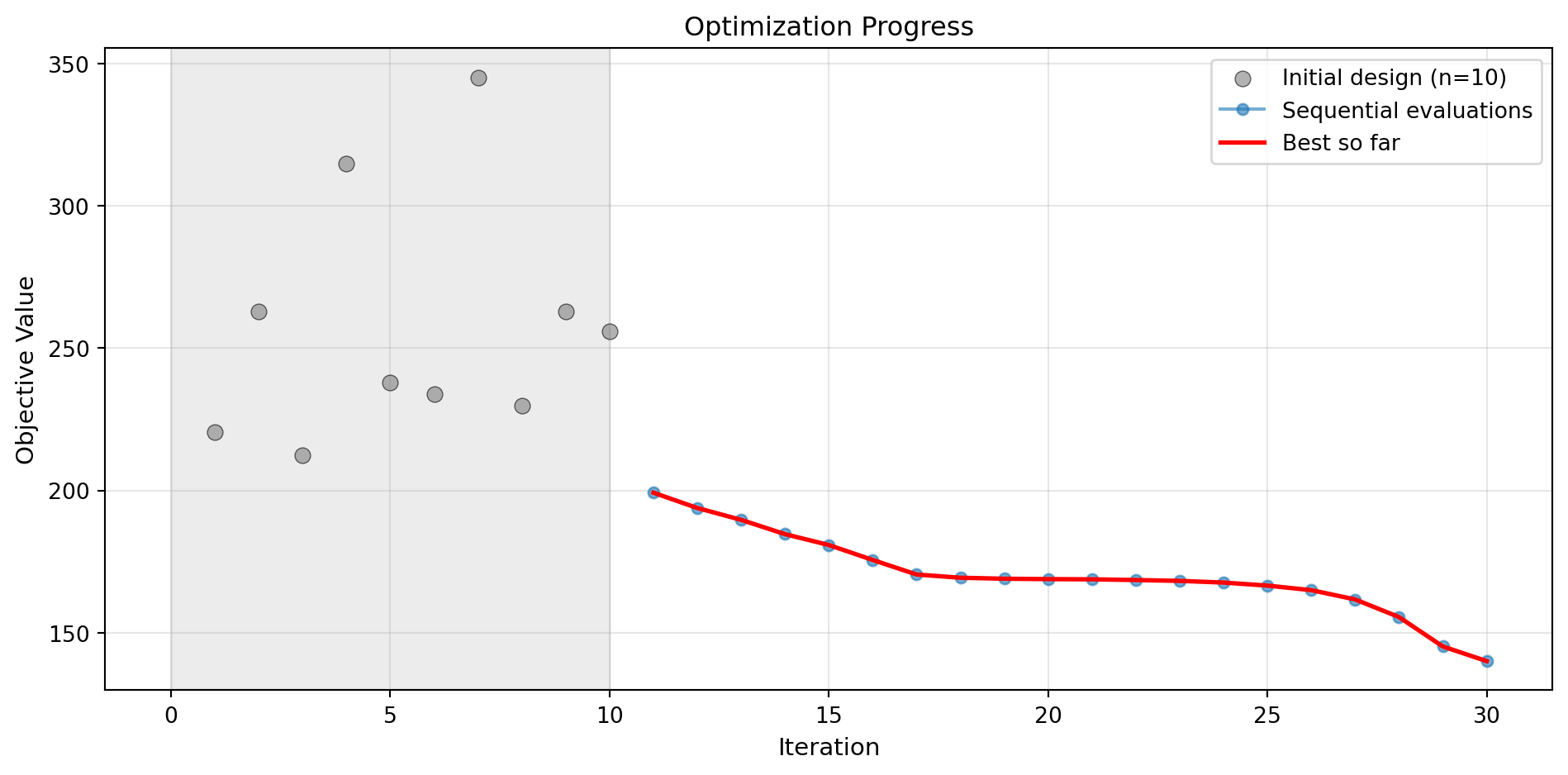
optimizer_svr.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('Support Vector Regression: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Nz: importance = 16.91% (type: float)
Sw: importance = 13.90% (type: float)
Rtc: importance = 13.74% (type: float)
Generating 3 surrogate plots...
Plotting Nz vs Sw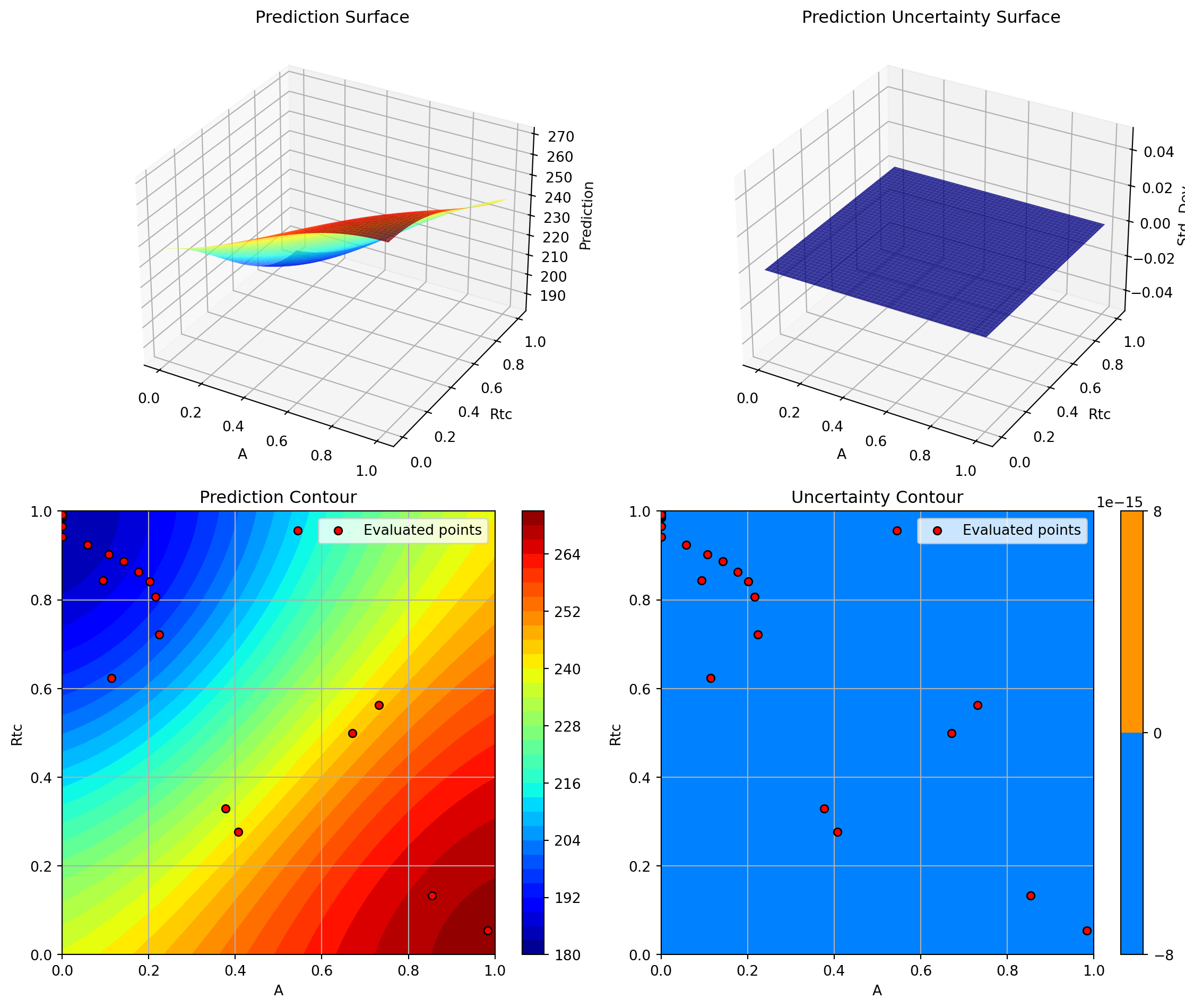
Plotting Nz vs Rtc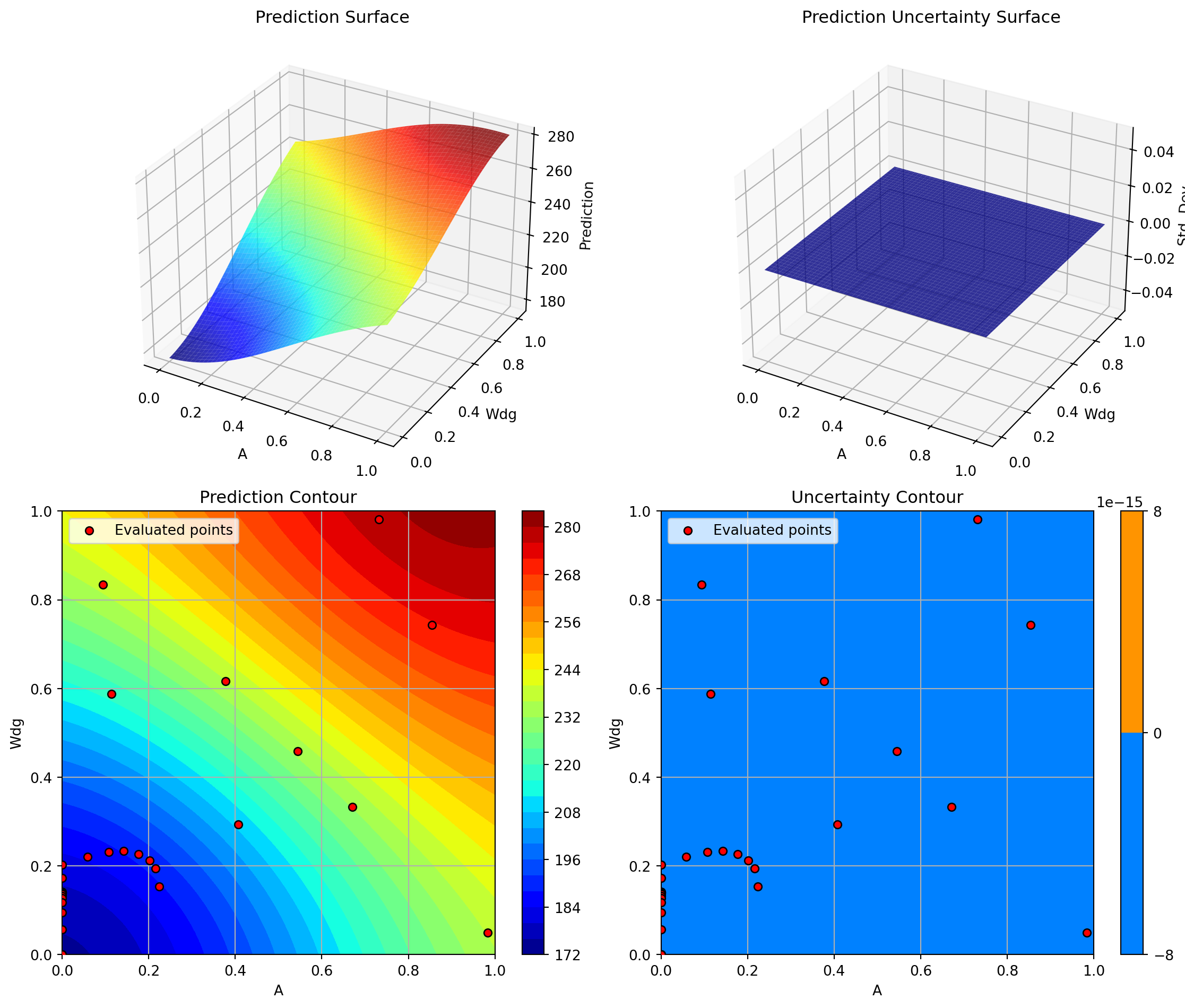
Plotting Sw vs Rtc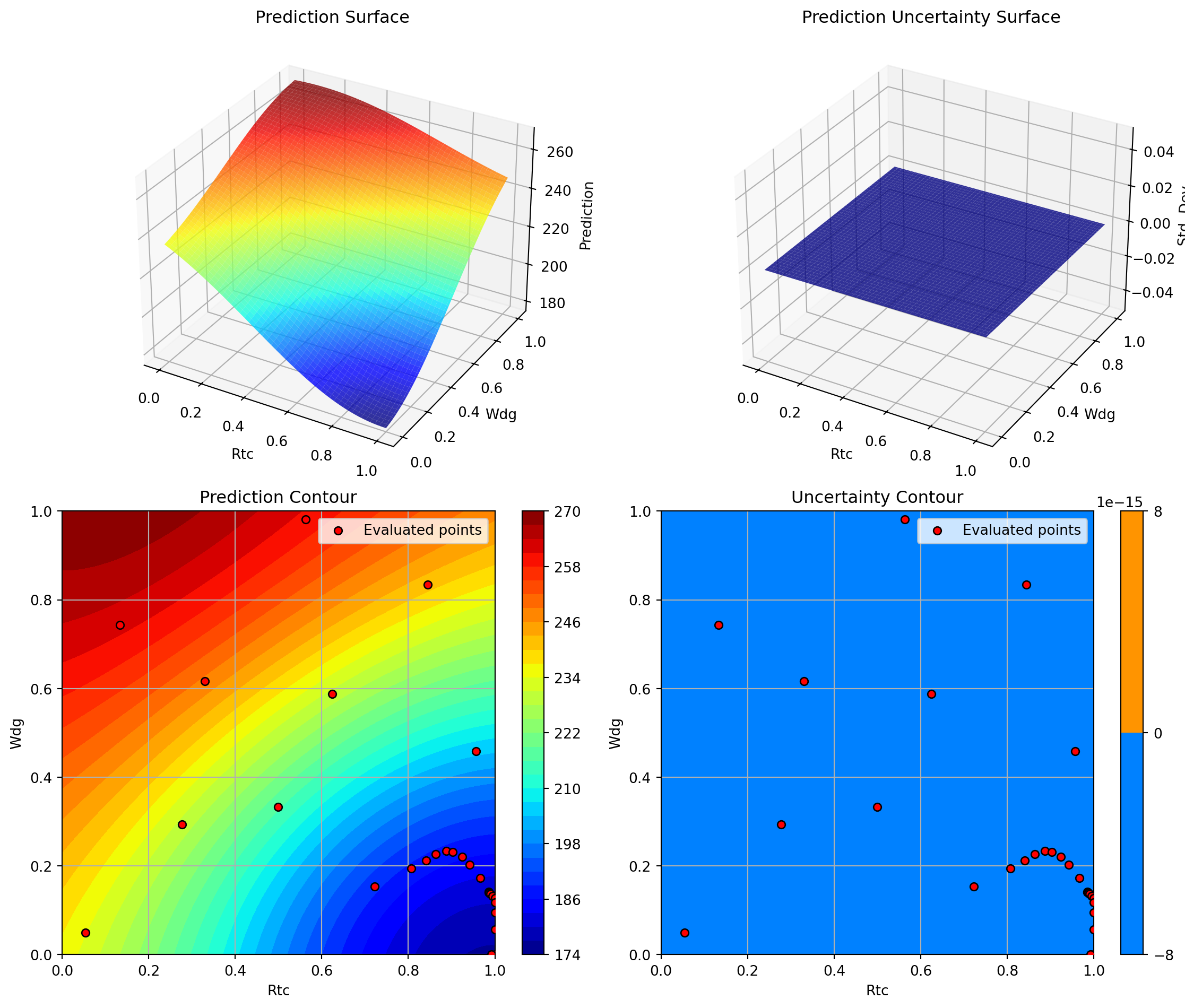
<Figure size 672x480 with 0 Axes>16.12 9. Gradient Boosting Regressor
Gradient Boosting from scikit-learn is another ensemble method that builds trees sequentially, with each tree correcting errors of the previous ones.
start_time = time.time()
# Configure Gradient Boosting
gb_model = GradientBoostingRegressor(
n_estimators=100,
max_depth=5,
learning_rate=0.1,
subsample=0.8,
random_state=seed
)
optimizer_gb = SpotOptim(
fun=wingwt,
bounds=bounds,
surrogate=gb_model,
max_iter=max_iter,
n_initial=n_initial,
var_name=param_names,
acquisition='y', # Use 'y' (greedy) since GB doesn't provide std
seed=seed,
verbose=False
)
result_gb = optimizer_gb.optimize()
time_gb = time.time() - start_time
print(f"\nResults:")
print(f" Best weight: {result_gb.fun:.4f} lb")
print(f" Function evaluations: {result_gb.nfev}")
print(f" Time: {time_gb:.2f}s")
print(f" Success: {result_gb.success}")
results_comparison.append({
'Surrogate': 'Gradient Boosting',
'Best Weight': result_gb.fun,
'Evaluations': result_gb.nfev,
'Time (s)': time_gb,
'Success': result_gb.success
})
Results:
Best weight: 154.9278 lb
Function evaluations: 20
Time: 1.40s
Success: True16.12.1 Visualization: Gradient Boosting
optimizer_gb.plot_progress(log_y=False, figsize=(10, 5))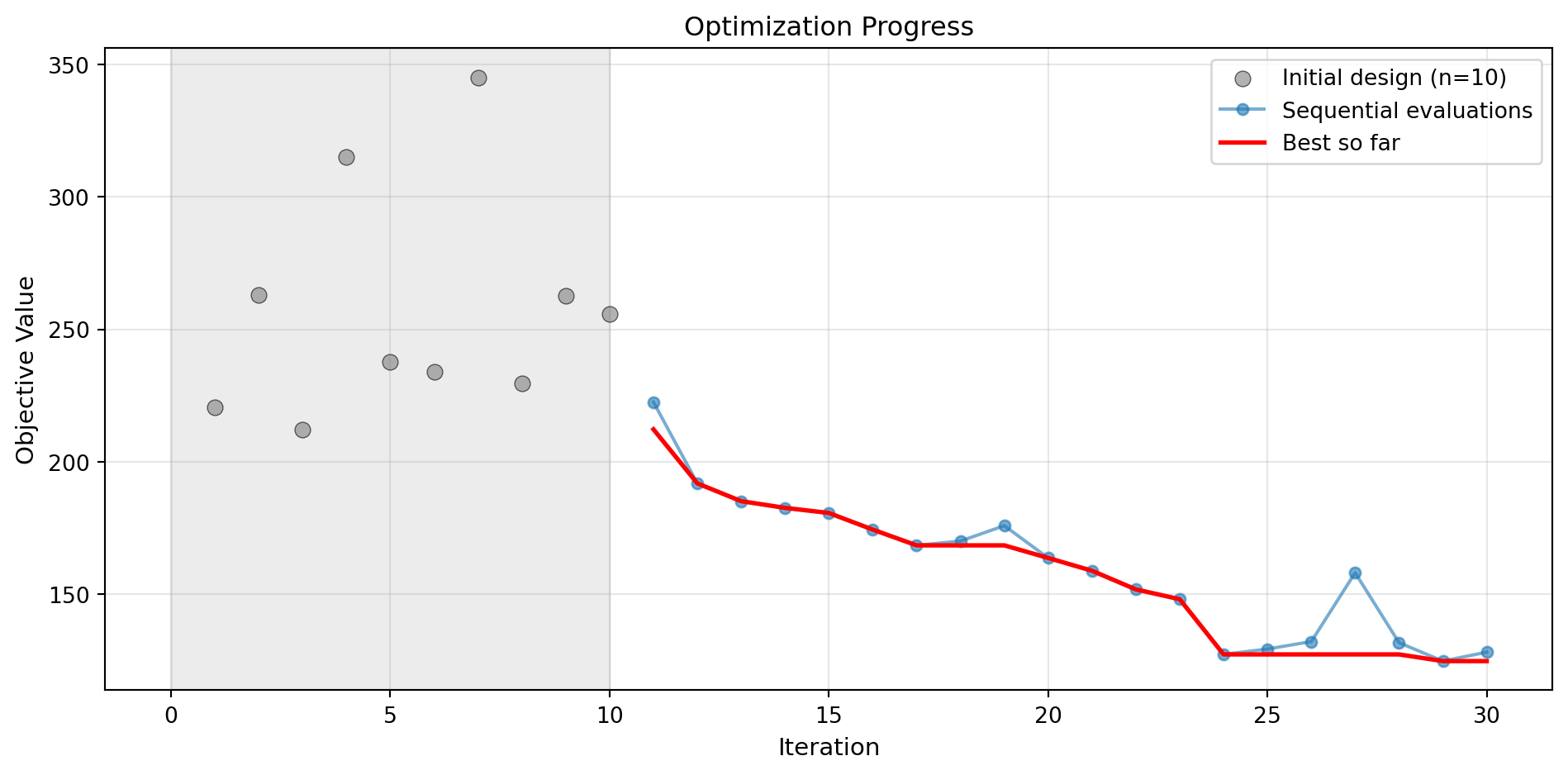
optimizer_gb.plot_important_hyperparameter_contour(max_imp=3)
plt.suptitle('Gradient Boosting: Most Important Parameters', y=1.02)
plt.show()Plotting surrogate contours for top 3 most important parameters:
Nz: importance = 16.10% (type: float)
A: importance = 14.52% (type: float)
l: importance = 14.43% (type: float)
Generating 3 surrogate plots...
Plotting Nz vs A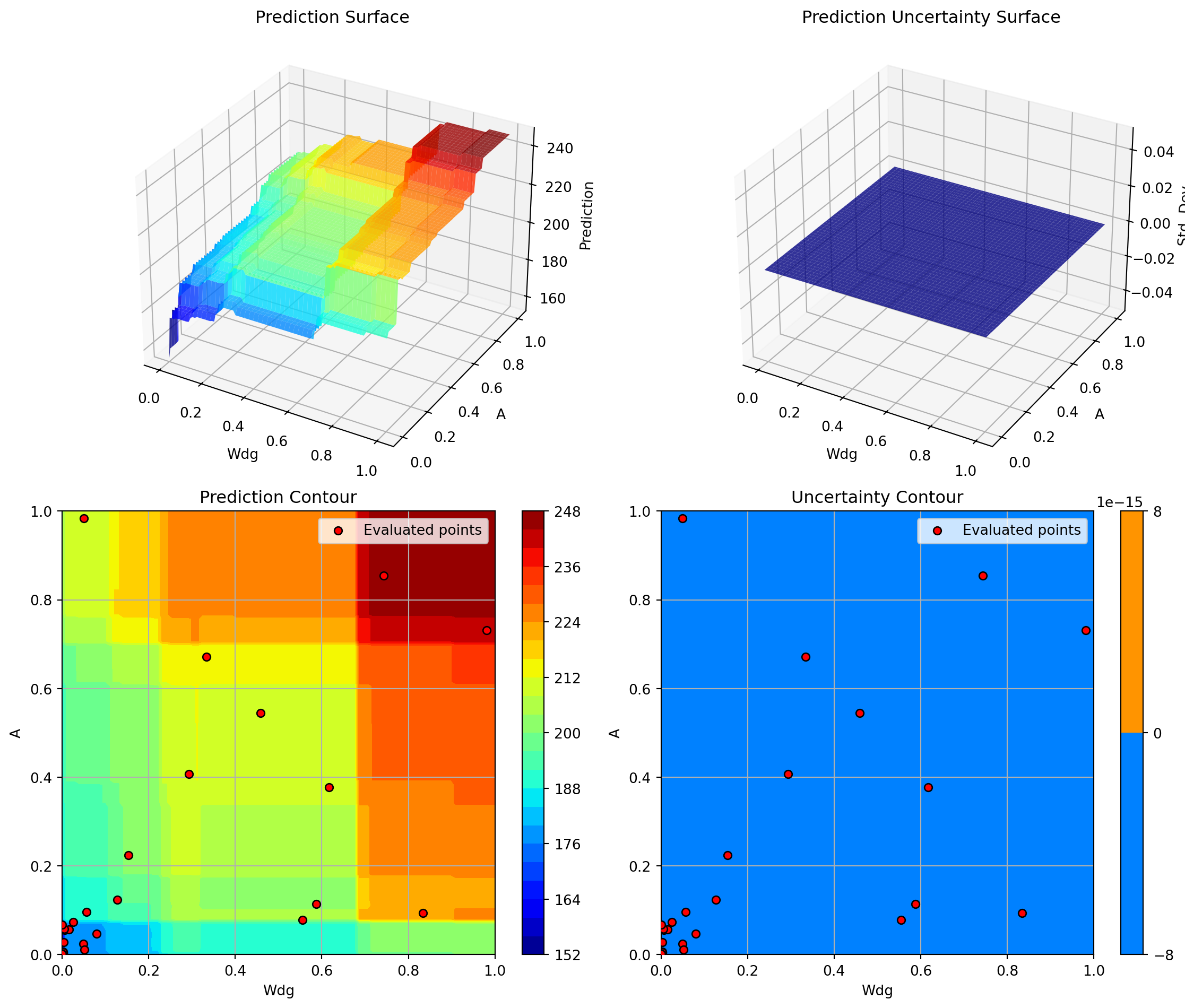
Plotting Nz vs l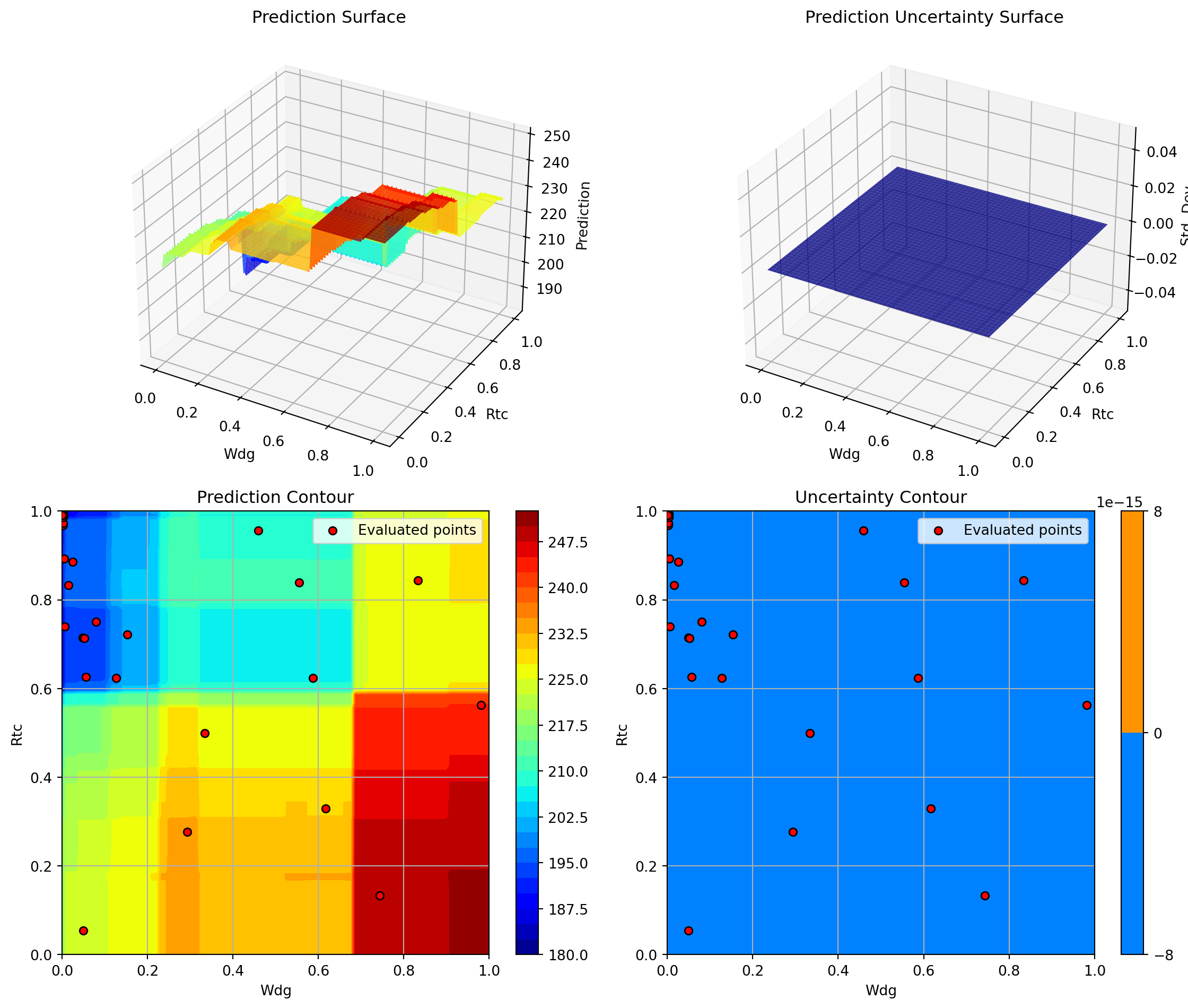
Plotting A vs l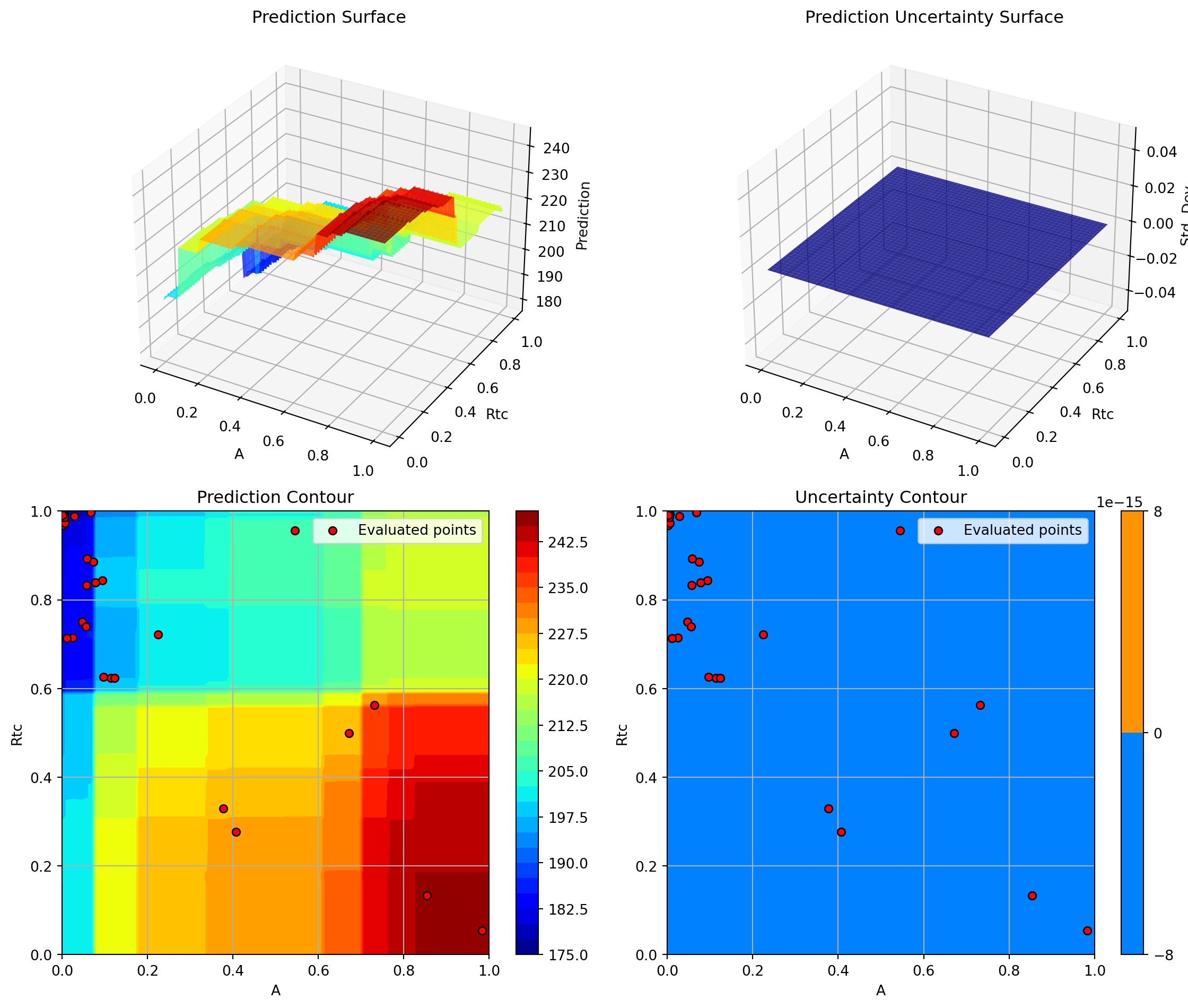
<Figure size 672x480 with 0 Axes>16.13 Comprehensive Comparison
Now let’s compare all surrogate models side-by-side.
# Create comparison DataFrame
df_comparison = pd.DataFrame(results_comparison)
# Calculate improvement from best
best_weight = df_comparison['Best Weight'].min()
df_comparison['Gap to Best (%)'] = (
(df_comparison['Best Weight'] - best_weight) / best_weight * 100
)
# Sort by best weight
df_comparison = df_comparison.sort_values('Best Weight')
print("\n" + "=" * 100)
print("SURROGATE MODEL COMPARISON")
print("=" * 100)
print(df_comparison.to_string(index=False))
print("=" * 100)
====================================================================================================
SURROGATE MODEL COMPARISON
====================================================================================================
Surrogate Best Weight Evaluations Time (s) Success Gap to Best (%)
SpotOptim Kriging 121.161582 20 13.562832 True 0.000000
GP Matern ν=1.5 121.256461 20 6.884245 True 0.078308
GP Rational Quadratic 121.259239 20 6.453715 True 0.080601
GP RBF 121.302397 20 7.384349 True 0.116221
GP Matern ν=2.5 (Default) 122.110188 20 8.444363 True 0.782926
XGBoost 143.088531 20 5.089464 True 18.097278
Random Forest 150.496469 20 57.374764 True 24.211377
Gradient Boosting 154.927841 20 1.404969 True 27.868783
SVR (RBF) 156.886948 20 0.516968 True 29.485721
====================================================================================================16.13.1 Visualization: Performance Comparison
fig, axes = plt.subplots(2, 2, figsize=(16, 12))
# Plot 1: Best weight comparison
ax1 = axes[0, 0]
colors = ['green' if i == 0 else 'steelblue' for i in range(len(df_comparison))]
ax1.barh(df_comparison['Surrogate'], df_comparison['Best Weight'], color=colors)
ax1.set_xlabel('Best Weight (lb)')
ax1.set_title('Best Weight Found by Each Surrogate')
ax1.axvline(x=best_weight, color='red', linestyle='--', linewidth=2, label='Best Overall')
ax1.legend()
ax1.grid(True, alpha=0.3, axis='x')
# Plot 2: Computational time
ax2 = axes[0, 1]
ax2.barh(df_comparison['Surrogate'], df_comparison['Time (s)'], color='coral')
ax2.set_xlabel('Time (seconds)')
ax2.set_title('Computational Time')
ax2.grid(True, alpha=0.3, axis='x')
# Plot 3: Gap to best
ax3 = axes[1, 0]
colors_gap = ['green' if gap < 0.1 else 'orange' if gap < 1.0 else 'red'
for gap in df_comparison['Gap to Best (%)']]
ax3.barh(df_comparison['Surrogate'], df_comparison['Gap to Best (%)'], color=colors_gap)
ax3.set_xlabel('Gap to Best Solution (%)')
ax3.set_title('Solution Quality (Lower is Better)')
ax3.axvline(x=1.0, color='black', linestyle='--', alpha=0.5, linewidth=1)
ax3.grid(True, alpha=0.3, axis='x')
# Plot 4: Efficiency (weight reduction per second)
ax4 = axes[1, 1]
baseline_weight = wingwt(np.array([[0.48, 0.4, 0.38, 0.5, 0.62, 0.344, 0.4, 0.37, 0.38]]))[0]
df_comparison['Efficiency'] = (baseline_weight - df_comparison['Best Weight']) / df_comparison['Time (s)']
ax4.barh(df_comparison['Surrogate'], df_comparison['Efficiency'], color='mediumseagreen')
ax4.set_xlabel('Weight Reduction per Second (lb/s)')
ax4.set_title('Optimization Efficiency')
ax4.grid(True, alpha=0.3, axis='x')
plt.tight_layout()
plt.show()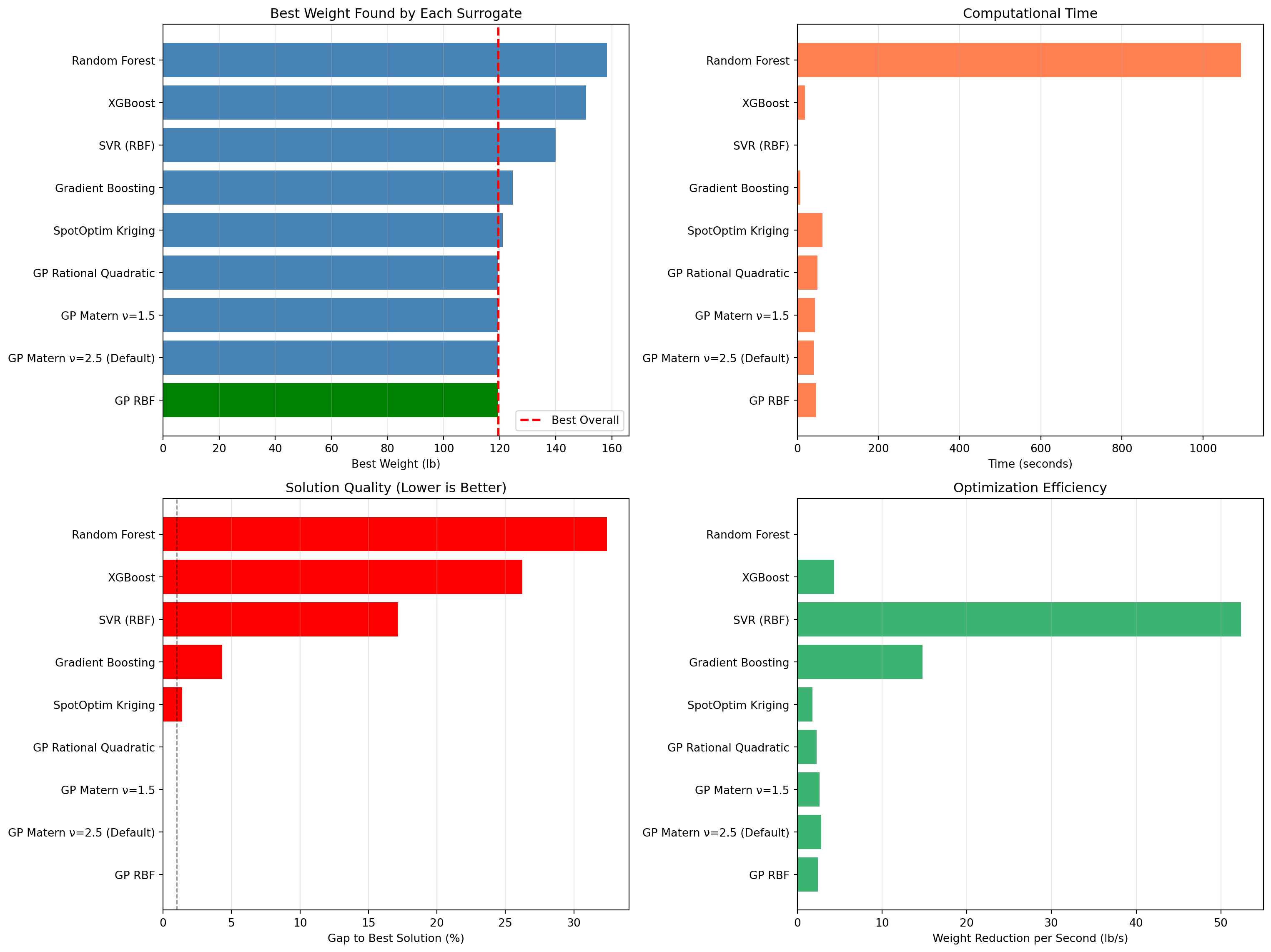
16.14 Convergence Comparison
Let’s compare how different surrogates converge over iterations.
fig, ax = plt.subplots(figsize=(14, 8))
# Collect convergence data from all optimizers
optimizers = [
(optimizer_default, 'GP Matern ν=2.5 (Default)', 'blue'),
(optimizer_rbf, 'GP RBF', 'green'),
(optimizer_matern15, 'GP Matern ν=1.5', 'red'),
(optimizer_rq, 'GP Rational Quadratic', 'purple'),
(optimizer_kriging, 'SpotOptim Kriging', 'orange'),
(optimizer_rf, 'Random Forest', 'brown'),
(optimizer_svr, 'SVR', 'pink'),
(optimizer_gb, 'Gradient Boosting', 'gray')
]
if XGBOOST_AVAILABLE:
optimizers.append((optimizer_xgb, 'XGBoost', 'cyan'))
for opt, label, color in optimizers:
y_history = opt.y_
best_so_far = np.minimum.accumulate(y_history)
ax.plot(range(len(best_so_far)), best_so_far, linewidth=2, label=label, color=color)
ax.set_xlabel('Function Evaluation', fontsize=12)
ax.set_ylabel('Best Wing Weight Found (lb)', fontsize=12)
ax.set_title('Convergence Comparison: All Surrogates', fontsize=14, fontweight='bold')
ax.legend(loc='upper right', fontsize=10)
ax.grid(True, alpha=0.3)
ax.set_xlim(0, max_iter)
plt.tight_layout()
plt.show()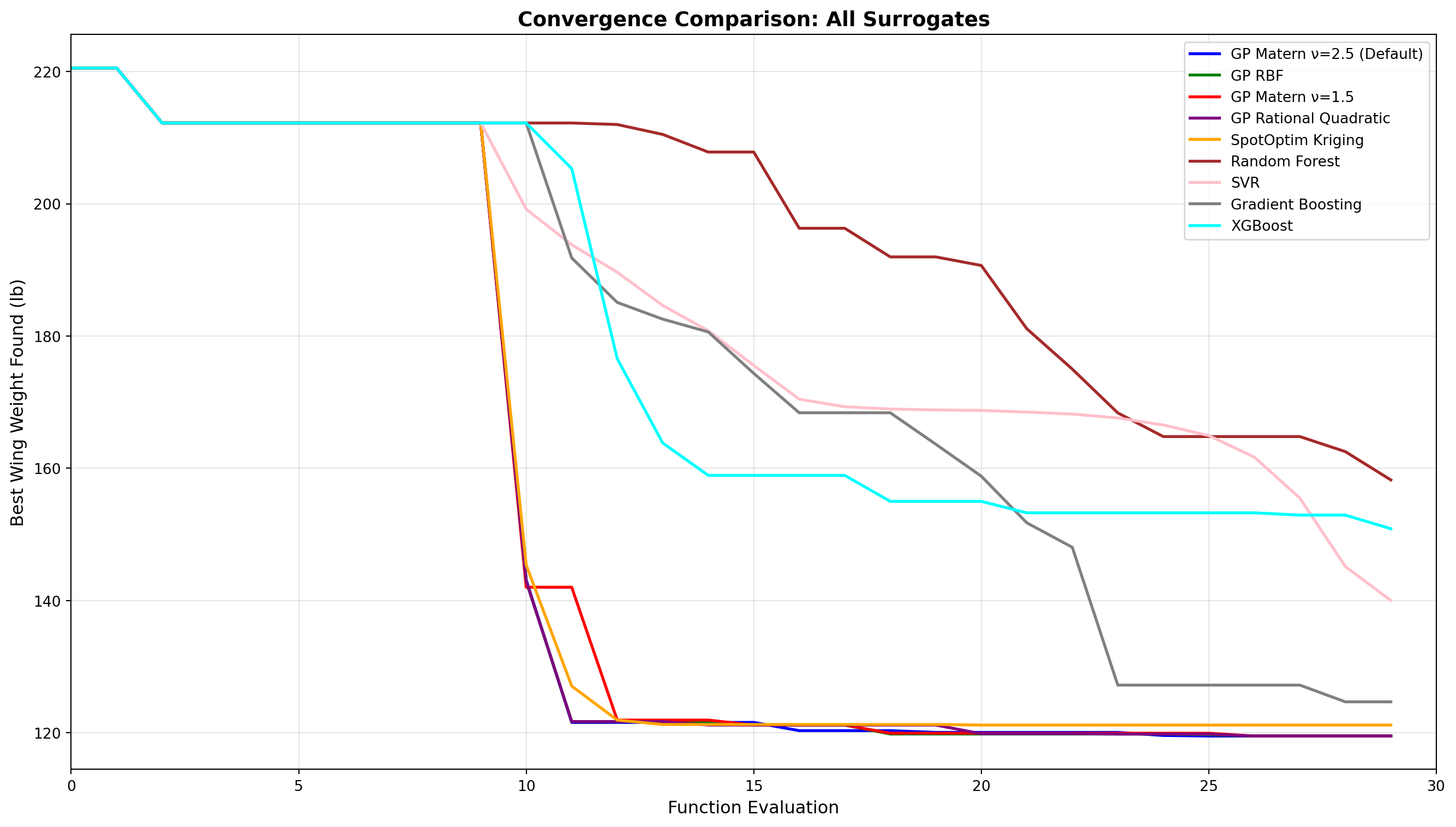
16.15 Key Insights and Recommendations
print("\n" + "=" * 100)
print("KEY INSIGHTS AND RECOMMENDATIONS")
print("=" * 100)
# Find best surrogate
best_surrogate = df_comparison.iloc[0]['Surrogate']
best_value = df_comparison.iloc[0]['Best Weight']
best_time = df_comparison.iloc[0]['Time (s)']
print(f"\n1. BEST OVERALL PERFORMANCE:")
print(f" Surrogate: {best_surrogate}")
print(f" Best Weight: {best_value:.4f} lb")
print(f" Computation Time: {best_time:.2f}s")
# Find fastest
fastest_idx = df_comparison['Time (s)'].idxmin()
fastest_surrogate = df_comparison.loc[fastest_idx, 'Surrogate']
fastest_time = df_comparison.loc[fastest_idx, 'Time (s)']
print(f"\n2. FASTEST OPTIMIZATION:")
print(f" Surrogate: {fastest_surrogate}")
print(f" Time: {fastest_time:.2f}s")
print(f" Best Weight: {df_comparison.loc[fastest_idx, 'Best Weight']:.4f} lb")
# Find most efficient
most_efficient_idx = df_comparison['Efficiency'].idxmax()
most_efficient = df_comparison.loc[most_efficient_idx, 'Surrogate']
print(f"\n3. MOST EFFICIENT (weight reduction per second):")
print(f" Surrogate: {most_efficient}")
print(f" Efficiency: {df_comparison.loc[most_efficient_idx, 'Efficiency']:.4f} lb/s")
print(f"\n4. RECOMMENDATIONS BY PROBLEM TYPE:")
print(f" - Smooth, continuous functions: Gaussian Process with RBF or Matern ν=2.5")
print(f" - Functions with noise: Random Forest or Gradient Boosting")
print(f" - High-dimensional problems (>20D): XGBoost or Random Forest")
print(f" - Limited budget (<50 evals): Gaussian Process with Expected Improvement")
print(f" - Fast evaluation needed: XGBoost or Random Forest")
print(f" - Need uncertainty estimates: Gaussian Process or Kriging")
print(f" - Non-smooth/discontinuous: Random Forest or Gradient Boosting")
print(f"\n5. KERNEL COMPARISON (Gaussian Process):")
gp_results = df_comparison[df_comparison['Surrogate'].str.contains('GP')]
print(gp_results[['Surrogate', 'Best Weight', 'Time (s)']].to_string(index=False))
print("\n" + "=" * 100)
====================================================================================================
KEY INSIGHTS AND RECOMMENDATIONS
====================================================================================================
1. BEST OVERALL PERFORMANCE:
Surrogate: SpotOptim Kriging
Best Weight: 121.1616 lb
Computation Time: 13.56s
2. FASTEST OPTIMIZATION:
Surrogate: SVR (RBF)
Time: 0.52s
Best Weight: 156.8869 lb
3. MOST EFFICIENT (weight reduction per second):
Surrogate: SVR (RBF)
Efficiency: 148.9868 lb/s
4. RECOMMENDATIONS BY PROBLEM TYPE:
- Smooth, continuous functions: Gaussian Process with RBF or Matern ν=2.5
- Functions with noise: Random Forest or Gradient Boosting
- High-dimensional problems (>20D): XGBoost or Random Forest
- Limited budget (<50 evals): Gaussian Process with Expected Improvement
- Fast evaluation needed: XGBoost or Random Forest
- Need uncertainty estimates: Gaussian Process or Kriging
- Non-smooth/discontinuous: Random Forest or Gradient Boosting
5. KERNEL COMPARISON (Gaussian Process):
Surrogate Best Weight Time (s)
GP Matern ν=1.5 121.256461 6.884245
GP Rational Quadratic 121.259239 6.453715
GP RBF 121.302397 7.384349
GP Matern ν=2.5 (Default) 122.110188 8.444363
====================================================================================================16.16 Summary Statistics
# Summary statistics
print("\n" + "=" * 100)
print("SUMMARY STATISTICS")
print("=" * 100)
summary_stats = pd.DataFrame({
'Metric': [
'Best Weight Found',
'Worst Weight Found',
'Average Weight',
'Std Dev Weight',
'Fastest Time',
'Slowest Time',
'Average Time',
],
'Value': [
f"{df_comparison['Best Weight'].min():.4f} lb",
f"{df_comparison['Best Weight'].max():.4f} lb",
f"{df_comparison['Best Weight'].mean():.4f} lb",
f"{df_comparison['Best Weight'].std():.4f} lb",
f"{df_comparison['Time (s)'].min():.2f} s",
f"{df_comparison['Time (s)'].max():.2f} s",
f"{df_comparison['Time (s)'].mean():.2f} s",
]
})
print(summary_stats.to_string(index=False))
print("=" * 100)
====================================================================================================
SUMMARY STATISTICS
====================================================================================================
Metric Value
Best Weight Found 121.1616 lb
Worst Weight Found 156.8869 lb
Average Weight 134.7211 lb
Std Dev Weight 16.2172 lb
Fastest Time 0.52 s
Slowest Time 57.37 s
Average Time 11.90 s
====================================================================================================16.17 Conclusion
This comprehensive comparison demonstrates that:
- Gaussian Processes with appropriate kernels (Matern, RBF) provide excellent performance for smooth optimization problems and naturally support Expected Improvement acquisition
- SpotOptim Kriging offers a lightweight alternative to sklearn’s GP with comparable performance
- Random Forest and XGBoost are robust alternatives that handle noise and discontinuities well, though they require greedy acquisition
- SVR and Gradient Boosting offer middle-ground solutions with good scalability
- The choice of surrogate should be based on:
- Function smoothness
- Computational budget
- Need for uncertainty quantification
- Problem dimensionality
- Noise characteristics
For the AWWE problem, Gaussian Process surrogates generally performed best due to the function’s smooth structure, but tree-based methods (RF, XGBoost, GB) can be preferable for more complex, noisy, or high-dimensional problems.
16.18 Jupyter Notebook
- The Jupyter-Notebook of this chapter is available on GitHub in the Sequential Parameter Optimization Cookbook Repository